GgaEX0008324 @ galGal4
Exon Skipping
Gene
ENSGALG00000005892 | SCUBE2
Description
signal peptide, CUB domain, EGF-like 2 [Source:HGNC Symbol;Acc:HGNC:30425]
Coordinates
chr5:8957480-8959958:+
Coord C1 exon
chr5:8957480-8957641
Coord A exon
chr5:8958937-8959134
Coord C2 exon
chr5:8959806-8959958
Length
198 bp
Sequences
Splice sites
3' ss Seq
ATACTGTGTTGTTTTTTCAGATA
3' ss Score
10.69
5' ss Seq
GCTGTAAGT
5' ss Score
8.56
Exon sequences
Seq C1 exon
TTCAGTGTTCGCCTGGTCATTTCTACAATACGACAACTCATCGGTGTATTCGCTGCATGGTTGGGACGTACCAGCCTGAGTTTGGACAGAATTACTGTATTTCTTGCCCTGGAAATACCACTACAGATTATGACGGATCCACAAATATAACGCAGTGCAAAA
Seq A exon
ATAGGCAGTGCGGAGGTGAACTGGGCGATTACACCGGGTATATTGAATCACCCAATTATCCTGGGGACTATCCTGCAAACACCGAATGCACTTGGAATATTAACCCACCGCCAAAGCGCCGCATTCTGATTGTGGTTCCAGAAATATTTCTACCAATAGAAGATGAGTGTGGGGACTACCTGGTGATGAGAAAAAGCT
Seq C2 exon
CTTCTTCCAACTCAGTGACCACGTATGAAACCTGTCAGACCTACGAACGCCCAATAGCTTTCACTTCCAGGTCTAAGAAGCTGTGGATCCAGTTCAAGTCAAATGAAGGGAATAGTGCCAAAGGATTCCAAGTTCCTTACGTAACATATGATG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000005892_CASSETTE3
Average complexity
S*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
Show structural model
Features
Disorder rate (Iupred):
C1=0.018 A=0.015 C2=0.000
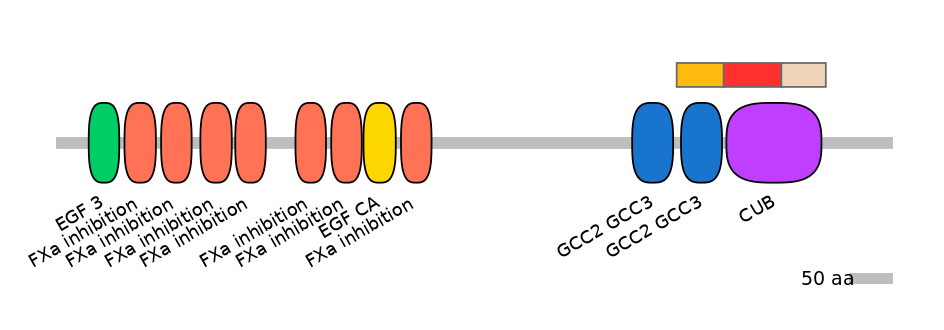
Domain overlap (PFAM):
C1:
PF076998=GCC2_GCC3=WD(100=87.3)
A:
PF0043115=CUB=PU(57.3=94.0)
C2:
PF0043115=CUB=PD(41.8=88.5)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TATTCGCTGCATGGTTGGGAC
R:
AGGAACTTGGAATCCTTTGGCA
Band lengths:
253-451
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]