HsaEX0002155 @ hg19
Exon Skipping
Gene
ENSG00000130402 | ACTN4
Description
actinin, alpha 4 [Source:HGNC Symbol;Acc:166]
Coordinates
chr19:39200897-39205201:+
Coord C1 exon
chr19:39200897-39200982
Coord A exon
chr19:39201916-39202001
Coord C2 exon
chr19:39205109-39205201
Length
86 bp
Sequences
Splice sites
3' ss Seq
TTCTCTCTCTCTCTGTGCAGATA
3' ss Score
13.12
5' ss Seq
AAGGTGAGC
5' ss Score
9.6
Exon sequences
Seq C1 exon
ACATCGTGAACACGGCCCGGCCCGACGAGAAGGCCATAATGACCTATGTGTCCAGCTTCTACCATGCCTTTTCAGGAGCGCAGAAG
Seq A exon
ATATTGTGGGCACTCTGAGGCCAGATGAGAAGGCCATCATGACTTACGTGTCCTGCTTCTACCACGCTTTCTCGGGGGCTCAGAAG
Seq C2 exon
GCTGAAACTGCCGCCAACCGGATCTGTAAGGTGCTGGCTGTCAACCAAGAGAACGAGCACCTGATGGAGGACTACGAGAAGCTGGCCAGCGAC
VastDB Features
Vast-tools module Information
Secondary ID
ENSG00000130402-'15-17,'15-15,16-17
Average complexity
C1
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
Show structural model
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF0030726=CH=PD(23.1=82.8)
A:
PF0030726=CH=PD(34.3=82.8),PF131021=Phage_int_SAM_5=PU(4.5=13.8)
C2:
PF131021=Phage_int_SAM_5=FE(34.1=100),PF0043516=Spectrin=PU(12.4=35.5),PF0103115=Dynamin_M=PU(21.1=48.4)
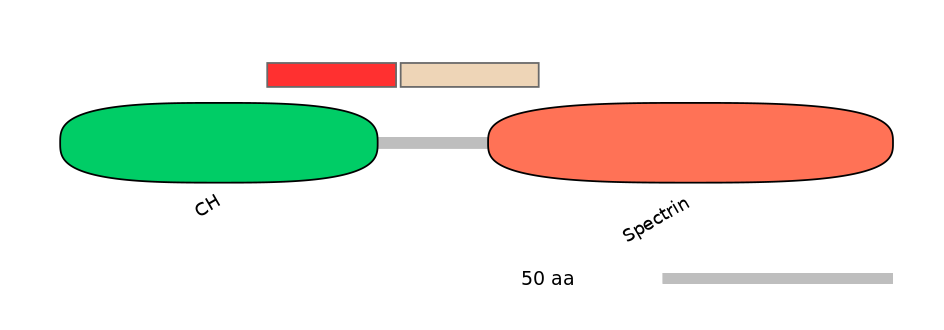
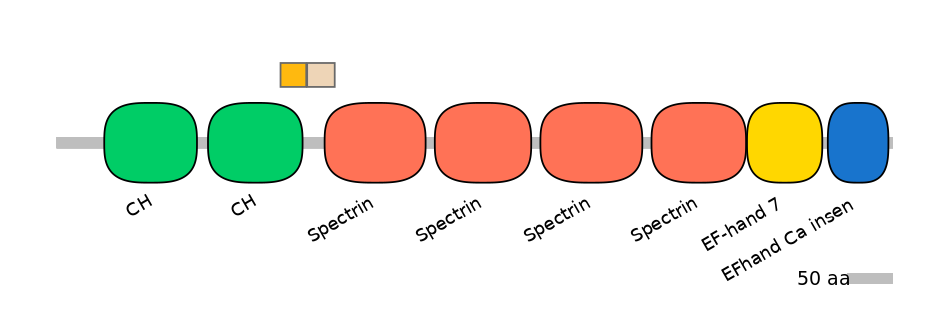
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
CCCGACGAGAAGGCCATAATG
R:
CCAGCTTCTCGTAGTCCTCCA
Band lengths:
151-237
Functional annotations
There are 2 annotated functions for this event
PMID: 15122314
This event
HsaEX0002154 and HsaEX0002155 are mutually exclusive. Expression of the splice variant (containing HsaEX0002155 instead of HsaEX0002154) was highly specific to SCLC cell lines (10/10), biopsies (3/3), and testis. The variant encoded a peptide with a three amino-acid change in exon 8, where the germline missense mutation takes place in familial focal segmental glomerulosclerosis (FSGS). The variant protein showed high affinity to filamentous actin polymers and was not localized with cortical actin. Alternatively spliced actinin-4 may be a new diagnostic marker of SCLC and a candidate target for selective therapy.
PMID: 22887464
This event
[Disease association only]. Variant actinin-4 (with Hsa0002155 instead of Hsa0002154) was expressed in 55% (96/176) of HGNTs, but in only 0.8% (3/378) of non-neuroendocrine (NE) lung cancers. The expression of variant actinin-4 was significantly associated with poorer overall survival in HGNT patients (P=0.00021, log-rank test). Multivariate analysis using the Cox proportional hazards model showed that the expression of variant actinin-4 was the most significant independent negative predictor of survival in HGNT patients (hazard ratio (HR), 2.15; P=0.00113) after the presence of lymph node metastasis (HR, 2.25; P=0.00023).
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- The Cancer Genome Atlas (TCGA)
- Genotype-Tissue Expression Project (GTEx)
- Autistic and control brains
- Pre-implantation embryo development
Other AS DBs:
FasterDB (Includes CLIP-seq data)
AS-ALPS (AS-induced ALteration of Protein Structure, links to PINs)
APPRIS (Selection of principal isoform)
DEU primates (Only for human)