RnoEX0006386 @ rn6
Exon Skipping
Gene
ENSRNOG00000020433 | Actn4
Description
actinin alpha 4 [Source:RGD Symbol;Acc:61816]
Coordinates
chr1:87092009-87097381:-
Coord C1 exon
chr1:87097300-87097381
Coord A exon
chr1:87095085-87095170
Coord C2 exon
chr1:87092009-87092101
Length
86 bp
Sequences
Splice sites
3' ss Seq
CTCTCTCTCTCTCTGCGCAGATA
3' ss Score
13.23
5' ss Seq
AAGGTGAGC
5' ss Score
9.6
Exon sequences
Seq C1 exon
GATGATCCAGTCACCAACCTAAACAATGCATTTGAAGTGGCTGAGAAATACCTTGATATCCCCAAGATGTTGGATGCTGAGG
Seq A exon
ATATTGTAGGCACTCTGAGGCCAGATGAGAAGGCCATCATGACTTACGTGTCCTGCTTCTACCACGCCTTCTCGGGGGCCCAGAAG
Seq C2 exon
GCTGAGACTGCTGCCAACCGGATCTGCAAGGTGCTGGCTGTGAACCAGGAAAATGAGCACCTGATGGAAGATTACGAGCGCCTGGCCAGTGAT
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000020433_MULTIEX1-2/2=C1-C2
Average complexity
ME(1-2[99=100])
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF0030726=CH=FE(26.0=100)
A:
PF0030726=CH=PD(23.1=82.8)
C2:
PF0043516=Spectrin=PU(9.9=35.5)
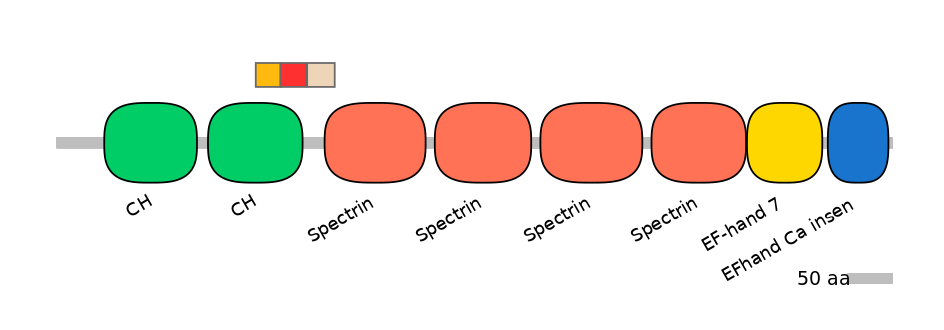
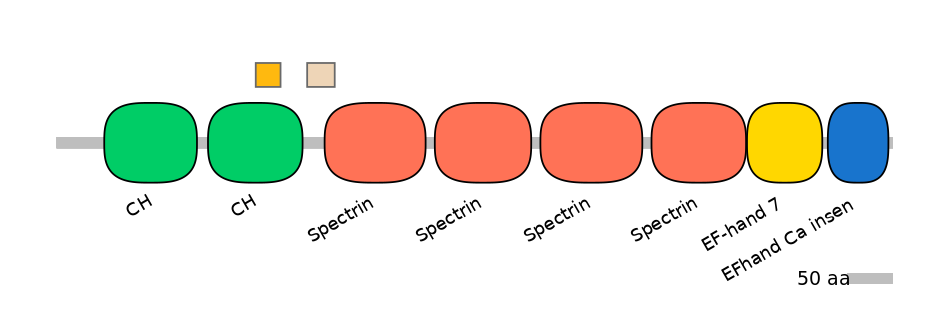
Other Inclusion Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
GATGATCCAGTCACCAACCTAAACA
R:
CCAGGCGCTCGTAATCTTCC
Band lengths:
167-253
Functional annotations
There are 2 annotated functions for this event
PMID: 15122314
HsaEX0002154 and HsaEX0002155 are mutually exclusive. Expression of the splice variant (containing HsaEX0002155 instead of HsaEX0002154) was highly specific to SCLC cell lines (10/10), biopsies (3/3), and testis. The variant encoded a peptide with a three amino-acid change in exon 8, where the germline missense mutation takes place in familial focal segmental glomerulosclerosis (FSGS). The variant protein showed high affinity to filamentous actin polymers and was not localized with cortical actin. Alternatively spliced actinin-4 may be a new diagnostic marker of SCLC and a candidate target for selective therapy.
PMID: 22887464
[Disease association only]. Variant actinin-4 (with Hsa0002155 instead of Hsa0002154) was expressed in 55% (96/176) of HGNTs, but in only 0.8% (3/378) of non-neuroendocrine (NE) lung cancers. The expression of variant actinin-4 was significantly associated with poorer overall survival in HGNT patients (P=0.00021, log-rank test). Multivariate analysis using the Cox proportional hazards model showed that the expression of variant actinin-4 was the most significant independent negative predictor of survival in HGNT patients (hazard ratio (HR), 2.15; P=0.00113) after the presence of lymph node metastasis (HR, 2.25; P=0.00023).
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]