RnoEX0078609 @ rn6
Exon Skipping
Gene
ENSRNOG00000005309 | Scn8a
Description
sodium voltage-gated channel alpha subunit 8 [Source:RGD Symbol;Acc:3638]
Coordinates
chr7:142661357-142672714:+
Coord C1 exon
chr7:142661357-142661530
Coord A exon
chr7:142666771-142666838
Coord C2 exon
chr7:142672430-142672714
Length
68 bp
Sequences
Splice sites
3' ss Seq
ATCTGTATTCTTTTCCATAGGTA
3' ss Score
11.11
5' ss Seq
AGGGTAAGG
5' ss Score
9.16
Exon sequences
Seq C1 exon
GCCTTTGAGGACATCTACATTGAGCAGAGGAAGACCATCCGCACCATCCTGGAGTATGCGGACAAGGTCTTCACCTACATCTTCATCCTGGAGATGTTGCTCAAGTGGACAGCCTACGGCTTCGTCAAGTTCTTCACCAATGCCTGGTGCTGGTTGGACTTCCTCATTGTGGCT
Seq A exon
GTACCATTAAGTTTGTCTGGCTTAATTTAATGGGAGCTTCTGGGAGCTGCAGACTGTAAAGGGCGAGG
Seq C2 exon
GTGGTGGTGAATGCCTTGGTGGGCGCCATCCCCTCCATCATGAATGTGCTGCTGGTGTGTCTCATCTTCTGGCTGATTTTCAGCATCATGGGAGTTAACCTGTTTGCGGGGAAATACCACTACTGCTTTAACGAGACTTCTGAAATCCGGTTCGAAATCGATATTGTCAACAATAAAACGGACTGTGAGAAGCTCATGGAGGGCAACAGCACGGAGATCCGATGGAAGAATGTCAAGATCAACTTTGACAATGTCGGAGCAGGGTACCTGGCCCTTCTTCAAGTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000005309_MULTIEX2-1/2=C1-C2
Average complexity
ME(1-2[100=100])
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence inclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF0052026=Ion_trans=PU(17.0=67.2)
A:
PF0052026=Ion_trans=PD(8.9=40.0)
C2:
PF0052026=Ion_trans=FE(40.9=100)
Main Inclusion Isoform:
ENSRNOT00000080923fB12996
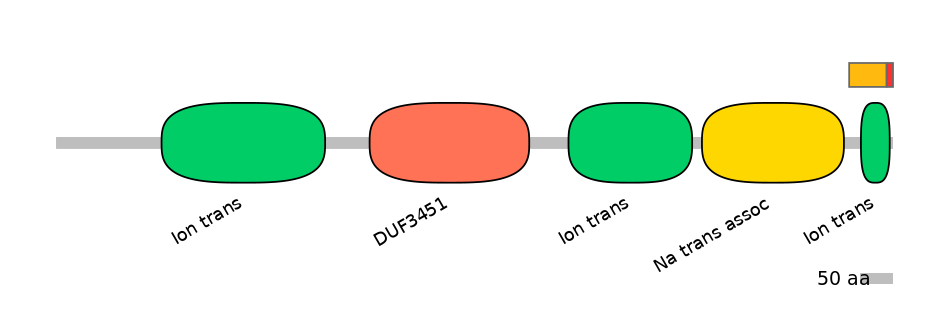
Other Inclusion Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGGAGTATGCGGACAAGGTCT
R:
ACACCAGCAGCACATTCATGA
Band lengths:
183-251
Functional annotations
There are 3 annotated functions for this event
PMID: 9295353
Mutually exclusive exons 18A (HsaEX0056569) and 18N (HsaEX0056568) determine whether a functional 18A+ Scn8a mRNA is produced. Inclusion of 18N introduces a premature termination codon and leads to nonsense-mediated decay, whereas skipping both 18A and 18N produces an isoform that lacks large portions of the third and fourth transmembrane domains.
PMID: 22434879
The 18N exon (neonatal form)(contains HsaEX0056568) has a stop codon in phase and although the mRNA can be detected by amplification methods, the truncated protein has not been observed. The switch from 18N to 18A (adult form)(contains HsaEX0056569) occurs only in a restricted set of neural tissues producing the functional channel while other tissues display the mRNA with the 18N exon also in adulthood. The mRNA species carrying the stop codon is subjected to Nonsense-Mediated Decay, providing a control mechanism of channel expression. The authors also map a string of cis-elements within the mutually exclusive exons and in the flanking introns responsible for their strict tissue and temporal specificity. These elements bind a series of positive (RbFox-1, SRSF1, SRSF2) and negative (hnRNPA1, PTB, hnRNPA2/B1, hnRNPD-like JKTBP) splicing regulatory proteins. These splicing factors, with the exception of RbFox-1, are ubiquitous but their levels vary during development and differentiation, ensuing unique sets of tissue and temporal levels of splicing factors.
PMID: 22044765
Transcripts containing exon 18N (HsaEX0056568) have a conserved in-frame stop codon that predicts the synthesis of a truncated, two-domain protein similar to the fetal form of the muscle calcium channel. They appear to be degraded by nonsense-mediated decay in HEK cells.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]