RnoEX0088089 @ rn6
Exon Skipping
Gene
ENSRNOG00000000483 | Syngap1
Description
synaptic Ras GTPase activating protein 1 [Source:RGD Symbol;Acc:621090]
Coordinates
chr20:5560152-5564437:+
Coord C1 exon
chr20:5560152-5560242
Coord A exon
chr20:5560989-5561068
Coord C2 exon
chr20:5563818-5564437
Length
80 bp
Sequences
Splice sites
3' ss Seq
CCCACTCTCTCCCCCTGCAGCTC
3' ss Score
12.54
5' ss Seq
CAGGTTGGT
5' ss Score
8.08
Exon sequences
Seq C1 exon
GCTGATGCTGGTGGAGGAGGAGCTGCGCCGGGACCACCCCGCCATGGCTGAGCCGCTGCCTGAACCCAAGAAGAGGCTGCTCGACGCTCAG
Seq A exon
CTCCTCATCAGGTAATTCTCCTGGTTCCGCTTTGACCACGGGCGGAGGACACTGGGGGAGGTGACTCCGGACCACTGCAG
Seq C2 exon
GAGAGGCAGCTTCCCCCCTTGGGTCCAACAAACCCGCGTGTGACGCTGGCCCCACCTTGGAACGGCCTGGCCCCCCCAGCCCCACCCCCCCCACCCCGGCTGCAGATCACAGAGAACGGCGAGTTCCGGAACACCGCAGACCACTAGCCCACCCAGCATCACAGACCTCCTTCCCTGTGCACCCTACCCCGGCCCACCCAGCGTCACAGACCTCCTTCCCAGTGCACCCGACCCTGGAACATCACCAACCACCAGGACTGGACGTCACCAAGGGACAGCGGGATTGTCTCCCTTAACGCCTCCTTGGGGCACCCATCTGTCAACCCCACTGCTCCATTCCAGGAGGGAGAGTGGGACCCTCAGCTGCCCTCTCACCCCAGGACACCACCTACCCCACACAGACCCCTTCACTCTGGGGTGCTATCCCCATCCTCTGCCTCACAGTTCCCCTGAGCACTGGGGGACAGACCCTCACCCCCACACTGGGGGTGTGGCACCTC
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000000483_CASSETTE2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref, Alt. Stop)
No structure available
Features
Disorder rate (Iupred):
C1=0.505 A=0.000 C2=NA
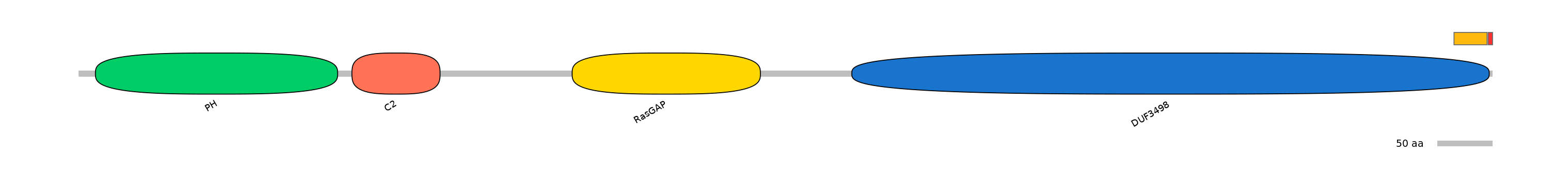
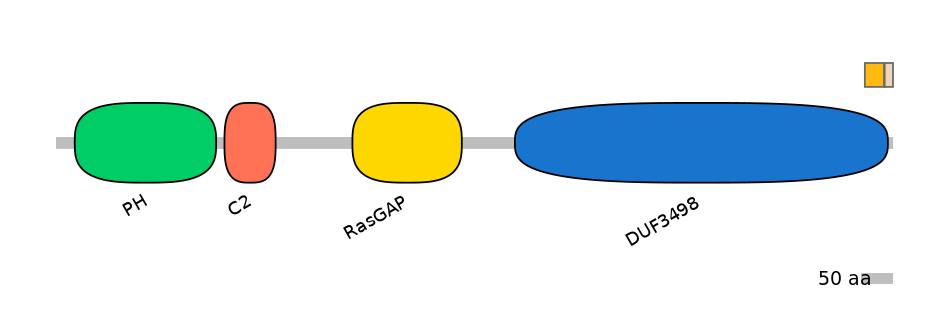
Domain overlap (PFAM):
C1:
PF120043=DUF3498=FE(5.2=100)
A:
PF120043=DUF3498=PD(0.2=20.0)
C2:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GAACCCAAGAAGAGGCTGCTC
R:
GCTAGTGGTCTGCGGTGTTC
Band lengths:
178-258
Functional annotations
There are 1 annotated functions for this event
PMID: 31911676
[CRISPR screen]. Conserved poison exon with pro-oncogenic impact when depleted: HeLa No Change at 14 days (FC=0.831, FDR=0.036), PC9 No Change at 14 days (FC=0.883, FDR=0.319), Late Xenograft Enriched (FC=1.949, FDR=0.000).
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]