BtaEX0031638 @ bosTau6
Exon Skipping
Gene
ENSBTAG00000020573 | SCUBE2
Description
signal peptide, CUB domain, EGF-like 2 [Source:HGNC Symbol;Acc:HGNC:30425]
Coordinates
chr15:44125660-44130950:+
Coord C1 exon
chr15:44125660-44125776
Coord A exon
chr15:44126017-44126103
Coord C2 exon
chr15:44130744-44130950
Length
87 bp
Sequences
Splice sites
3' ss Seq
ACCCACCTTCTGTCTGTCAGGAC
3' ss Score
8.37
5' ss Seq
GGGGTAATT
5' ss Score
5.8
Exon sequences
Seq C1 exon
AAGTGAAGGGGGGCCTGCCCACTAGCCTGTCACCCCACGTGTCCCTGCAGTGCGGTAAAAGTGGTGGAGGAGACAGGTGCTTCCTCAGATGTCACTCTGGCATTCACCTCTCTCCAG
Seq A exon
GACTGCAAGAGGCCTATTCTGTCACCTGCGGCTCTTCCTCTCCTCTCAGGAACAAACAGCAAAAATCAAATGACTCCGCTTTCGGGG
Seq C2 exon
TGTCTTGTGATCTCAGTTGCATTGTAAAGCGAACAGAGAAGCGGCTCCGGAAGGCCATCCGCACTCTCCGAAAGGCTGCCCACAGGGAGCAGCTGCACCTCCAGCTCTCAGGCGTGAACCTTGAAGTGGCTAAAAAGTCTCCCCGAACATCTGAACGCCAGATAGAGTCCTGTGCAATGGGCCAGGACCATGGAGGAAACCAGTGTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000020573_MULTIEX2-4/8=3-7
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.267 C2=0.248
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF075629=NCD3G=PU(0.1=0.0)
Main Inclusion Isoform:
ENSBTAT00000027416fB6177
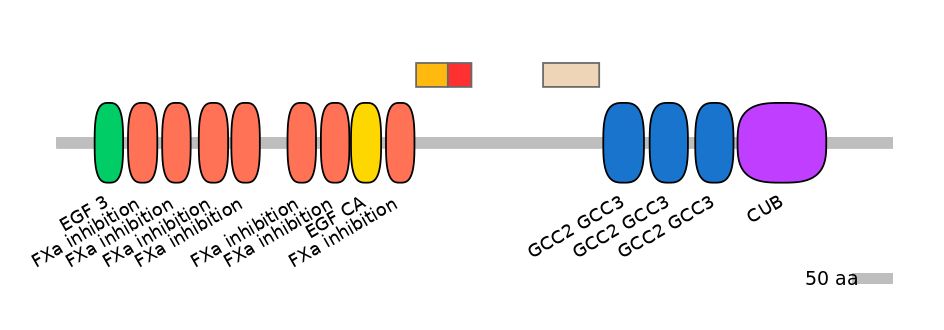
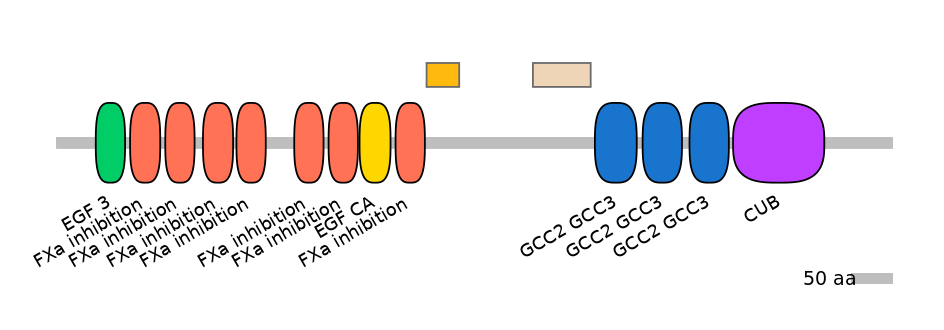
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GGAGACAGGTGCTTCCTCAGA
R:
TAGCCACTTCAAGGTTCACGC
Band lengths:
181-268
Functional annotations
There are 1 annotated functions for this event
PMID: 32249828
[CRISPR screen]. This alternative exon is implicated in cell fitness by a CRISPR-based exon deletion phenotypic screen in RPE-1 cells.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Pre-implantation embryo development