GgaEX0025048 @ galGal3
Exon Skipping
Gene
ENSGALG00000016890 | A3F958_CHICK
Description
NA
Coordinates
chr1:149566057-149571146:-
Coord C1 exon
chr1:149571066-149571146
Coord A exon
chr1:149566861-149566914
Coord C2 exon
chr1:149566057-149566210
Length
54 bp
Sequences
Splice sites
3' ss Seq
TCGCTGCCTGTTATTAAAAGACT
3' ss Score
2.9
5' ss Seq
CTGGTACTT
5' ss Score
4.74
Exon sequences
Seq C1 exon
GCCAAAATCAAAGCGGCGCAACATCAAGCTAACCAGGCTGCAGTGGCAGCCCAGGCAGCTGCGGCAGCTGCGACTGTGATG
Seq A exon
ACTCAGTCGACTGCCAAAGCAATGAAGCGACCTCTCGAAGCAACTGTAGACCTG
Seq C2 exon
GCCTTTCCTCCCGGTGTTCTTCACCCTTTACCAAAGAGACAAGCACTTGAAAAAAGCAATGGTGCCAGTGCCGTTTTCAATCCCAGTGTCTTGCACTATCAACAGGCTCTTGCCAATGCTCAGTTGCAGCAACATGCAGCGTTCATCCCGACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000016890_MULTIEX2-4/4=3-C2
Average complexity
S
Mappability confidence:
NA
Protein Impact
Alternative protein isoforms (Ref)
Show structural model
Features
Disorder rate (Iupred):
C1=0.136 A=0.167 C2=0.141
Domain overlap (PFAM):
C1:
PF0064219=zf-CCCH=PD(81.5=25.0)
A:
NO
C2:
NO
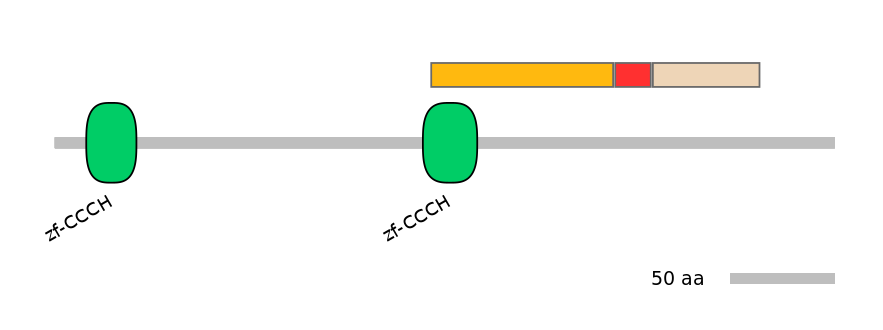
Main Skipping Isoform:
NA
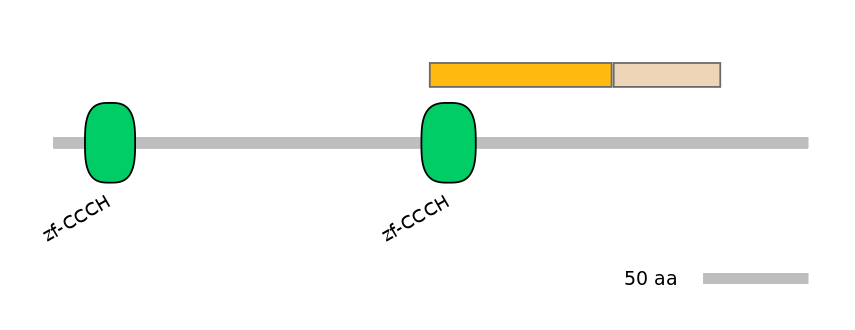
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
ACATCAAGCTAACCAGGCTGC
R:
GATTGAAAACGGCACTGGCAC
Band lengths:
143-197
Functional annotations
There are 2 annotated functions for this event
PMID: 21454535
Inferred from MBNL1: The exon 5 and 6 regions are both needed to control the nuclear localization of MBNL1
PMID: 27733504
The presence of alternative ex.54nt always significantly elevated MBNL1 and MBNL2 protein levels (_2.7 and _4.0-times, respectively). There were marginal changes of the GFP-MBNL level for constructs carrying ex.36nt (_1.3-times) and ex.95nt (_0.8-times). There were no significant differences between the activities of the analyzed proteins for the majority (67%) of AS events. However, these exons might have either a positive or negative effect on some specific AS events (Figure 5A and C). Interestingly, exOFF events predominated in AS events with a negative effect of ex.36nt (95%; P = 0.002) and a positive effect of ex.95nt (90%; P = 0.010). All together, these data indicate that the presence of sequences encoded by these alternative exons can significantly modulate MBNL splicing activity, but this regulation depends strongly on the targeted RNA.