GgaEX0025048 @ galGal4
Exon Skipping
Gene
ENSGALG00000016890 | MBNL2
Description
muscleblind-like 2 isoform 5 [Source:RefSeq peptide;Acc:NP_001156846]
Coordinates
chr1:144597391-144602663:-
Coord C1 exon
chr1:144602400-144602663
Coord A exon
chr1:144598195-144598248
Coord C2 exon
chr1:144597391-144597544
Length
54 bp
Sequences
Splice sites
3' ss Seq
TCGCTGCCTGTTATTAAAAGACT
3' ss Score
2.9
5' ss Seq
CTGGTACTT
5' ss Score
4.74
Exon sequences
Seq C1 exon
GTGTGCAGGGAGTTCCAGAGGGGAAACTGTGCGCGTGGAGAGACTGATTGCCGCTTTGCACATCCGGCAGACAGCACAATGATTGACACAAACGACAACACCGTAACCGTGTGTATGGATTACATAAAAGGTCGTTGCATGAGGGAGAAATGCAAGTATTTTCACCCTCCTGCACATTTGCAGGCCAAAATCAAAGCGGCGCAACATCAAGCTAACCAGGCTGCAGTGGCAGCCCAGGCAGCTGCGGCAGCTGCGACTGTGATG
Seq A exon
ACTCAGTCGACTGCCAAAGCAATGAAGCGACCTCTCGAAGCAACTGTAGACCTG
Seq C2 exon
GCCTTTCCTCCCGGTGTTCTTCACCCTTTACCAAAGAGACAAGCACTTGAAAAAAGCAATGGTGCCAGTGCCGTTTTCAATCCCAGTGTCTTGCACTATCAACAGGCTCTTGCCAATGCTCAGTTGCAGCAACATGCAGCGTTCATCCCGACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000016890_MULTIEX2-4/4=3-C2
Average complexity
S
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
Show structural model
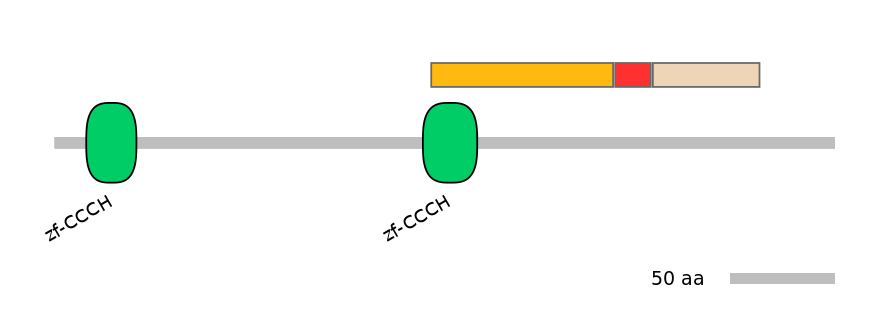
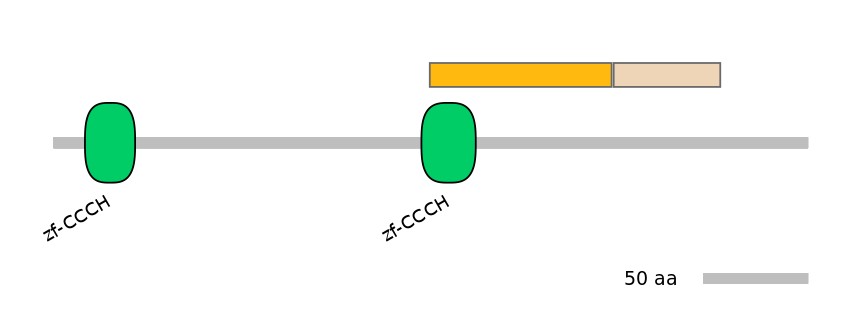
Features
Disorder rate (Iupred):
C1=0.148 A=0.236 C2=0.157
Domain overlap (PFAM):
C1:
PF0064219=zf-CCCH=PD(81.5=25.0)
A:
PF0273412=Dak2=PD(18.0=88.9)
C2:
NO
Associated events
Other assemblies
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
ACATCAAGCTAACCAGGCTGC
R:
GATTGAAAACGGCACTGGCAC
Band lengths:
143-197
Functional annotations
There are 2 annotated functions for this event
PMID: 21454535
Inferred from MBNL1: The exon 5 and 6 regions are both needed to control the nuclear localization of MBNL1
PMID: 27733504
The presence of alternative ex.54nt always significantly elevated MBNL1 and MBNL2 protein levels (_2.7 and _4.0-times, respectively). There were marginal changes of the GFP-MBNL level for constructs carrying ex.36nt (_1.3-times) and ex.95nt (_0.8-times). There were no significant differences between the activities of the analyzed proteins for the majority (67%) of AS events. However, these exons might have either a positive or negative effect on some specific AS events (Figure 5A and C). Interestingly, exOFF events predominated in AS events with a negative effect of ex.36nt (95%; P = 0.002) and a positive effect of ex.95nt (90%; P = 0.010). All together, these data indicate that the presence of sequences encoded by these alternative exons can significantly modulate MBNL splicing activity, but this regulation depends strongly on the targeted RNA.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]