DmeEX0008587 @ dm6
Exon Skipping
Gene
FBgn0265487 | mbl
Description
The gene muscleblind is referred to in FlyBase by the symbol Dmelmbl (CG33197, FBgn0265487). It is a protein_coding_gene from Dmel. It has 19 annotated transcripts and 19 polypeptides (18 unique). Gene sequence location is 2R:17216549..17379376. Its molecular function is described by: RNA binding; metal ion binding; nucleic acid binding. It is involved in the biological process described with 9 unique terms, many of which group under: regulation of nucleobase-containing compound metabolic process; reproductive process; programmed cell death; apoptotic process; organic cyclic compound metabolic process. 93 alleles are reported. The phenotypes of these alleles manifest in: fascicle; larva; rhabdomere; visceral muscle; cytoskeleton. The phenotypic classes of alleles include: increased mortality during development; some die during embryonic stage; phenotype; courtship behavior defective.
Coordinates
chr2R:17352129-17362097:+
Coord C1 exon
chr2R:17352129-17352305
Coord A exon
chr2R:17361113-17361154
Coord C2 exon
chr2R:17361980-17362097
Length
42 bp
Sequences
Splice sites
3' ss Seq
ATCGATAAATGCCAATAAAGATG
3' ss Score
-3.18
5' ss Seq
TTCGTAAGT
5' ss Score
9.77
Exon sequences
Seq C1 exon
GCAACAAACCCCTATCTGACCGGCATTCCGGCCAACTCGTACAGTCCGTACTACACAACGGGACACCTGGTGCCCGCCTTGCTGGGTCCGGATCCCGTGACCTCCCAACTGGGACCCGTGGTGCCCCAAACAGTGCAGGTGGCACAGCAGAAAATACCACGTTCCGACAGATTAGAG
Seq A exon
ATGGATGTTAAAACGGTTGGTTCATTTTACTATGACAACTTC
Seq C2 exon
CAATTCTCTGGCATGGTACCGTTCAAACGTCCAGCTGCCGAAAAGTCTGGCATTCCAGTTTATCAGCCCGGTGCGACCGCCTATCAGCAGCTAATGCAGCCCTACGTGCCAGTCTCAT
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0265487-'18-23,'18-22,24-23=AN
Average complexity
A_C2
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.299 A=0.000 C2=0.420
Domain overlap (PFAM):
C1:
PF0064219=zf-CCCH=PU(11.1=5.1)
A:
NO
C2:
NO
Main Inclusion Isoform:
FBpp0297559
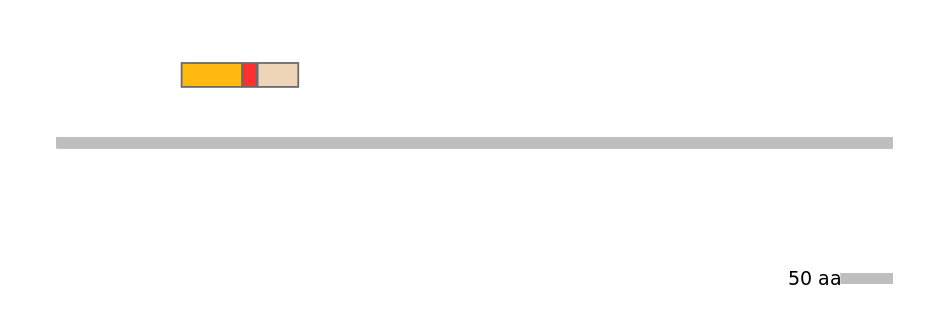
Main Skipping Isoform:
FBpp0297558
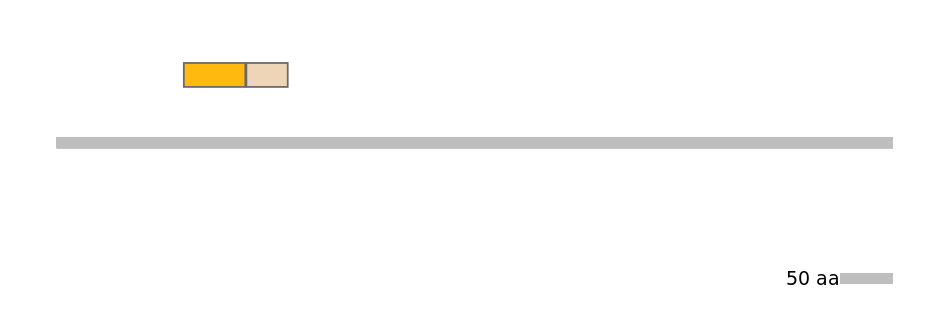
Other Inclusion Isoforms:
FBpp0086144, FBpp0086146, FBpp0307871, FBpp0307873
Other Skipping Isoforms:
FBpp0297556, FBpp0297561, FBpp0301759, FBpp0301760, FBpp0302029, FBpp0307872
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
GGCACAGCAGAAAATACCACG
R:
CATTAGCTGCTGATAGGCGGT
Band lengths:
133-175
Functional annotations
There are 7 annotated functions for this event
PMID: 21454535
The exon 5 and 6 regions are both needed to control the nuclear localization of MBNL1
PMID: 27733504
The presence of alternative ex.54nt always significantly elevated MBNL1 and MBNL2 protein levels (_2.7 and _4.0-times, respectively). There were marginal changes of the GFP-MBNL level for constructs carrying ex.36nt (_1.3-times) and ex.95nt (_0.8-times). There were no significant differences between the activities of the analyzed proteins for the majority (67%) of AS events. However, these exons might have either a positive or negative effect on some specific AS events (Figure 5A and C). Interestingly, exOFF events predominated in AS events with a negative effect of ex.36nt (95%; P = 0.002) and a positive effect of ex.95nt (90%; P = 0.010). All together, these data indicate that the presence of sequences encoded by these alternative exons can significantly modulate MBNL splicing activity, but this regulation depends strongly on the targeted RNA.
PMID: 29955876
Inclusion/exclusion of the exon affects splicing activity with sequential and overlapping binding motifs, as shown by competition assays between isoforms.
PMID: 31911676
Nuclear localization. Induction of MBNL1 exon skipping drove expected functional consequences. Nuclear levels of total MBNL1 were quantitatively lower following delivery of each pgRNA that induced appreciable exon skipping.
PMID: 21454535
Inferred from MBNL1: The exon 5 and 6 regions are both needed to control the nuclear localization of MBNL1
PMID: 27733504
The presence of alternative ex.54nt always significantly elevated MBNL1 and MBNL2 protein levels (_2.7 and _4.0-times, respectively). There were marginal changes of the GFP-MBNL level for constructs carrying ex.36nt (_1.3-times) and ex.95nt (_0.8-times). There were no significant differences between the activities of the analyzed proteins for the majority (67%) of AS events. However, these exons might have either a positive or negative effect on some specific AS events (Figure 5A and C). Interestingly, exOFF events predominated in AS events with a negative effect of ex.36nt (95%; P = 0.002) and a positive effect of ex.95nt (90%; P = 0.010). All together, these data indicate that the presence of sequences encoded by these alternative exons can significantly modulate MBNL splicing activity, but this regulation depends strongly on the targeted RNA.
PMID: 31730826
In this study, the authors first identified the discriminative expression and splicing profiles of the muscleblind-like 1 (MBNL1) gene in postnatal brown adipose tissues (BATs) compared to those of embryonic BATs. A shift in the MBNL1+ex 5 isoform 7 (MBNL1-7) to MBNL1-ex 5 isoform 1 (MBNL1-1) was characterized throughout BAT development or during the in vitro browning of pre-WAs, 3T3-L1 cells. The interplay between MBNL1 and the exonic CCUG motif constitutes an autoregulatory mechanism for excluding MBNL1 exon 5. The simultaneous association of RNA-binding motif protein 4a (RBM4a) with exonic and intronic CU elements collaboratively mediates the skipping of MBNL1 exon 5. Overexpressing the MBNL1-1 isoform exhibited a more-prominent effect than that of the MBNL1-7 isoform on programming its own transcripts and beige cell-related splicing events in a CCUG motif-mediated manner. In addition to splicing regulation, overexpression of the MBNL1-1 and MBNL1-7 isoforms differentially enhanced beige adipogenic signatures of 3T3-L1 cells.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)