RnoEX0078614 @ rn6
Exon Skipping
Gene
ENSRNOG00000006639 | Scn9a
Description
sodium voltage-gated channel alpha subunit 9 [Source:RGD Symbol;Acc:69368]
Coordinates
chr3:52656582-52659272:-
Coord C1 exon
chr3:52659144-52659272
Coord A exon
chr3:52657770-52657861
Coord C2 exon
chr3:52656582-52656794
Length
92 bp
Sequences
Splice sites
3' ss Seq
ACTCTGATTTAATCCTACAGGTA
3' ss Score
9.61
5' ss Seq
CAGGTAAGA
5' ss Score
10.77
Exon sequences
Seq C1 exon
GTACACTTTTACTGGGATATATACTTTTGAATCACTCATAAAAATCCTTGCAAGAGGCTTTTGCGTGGGAGAATTCACCTTCCTCCGTGACCCTTGGAACTGGCTGGACTTTGTTGTCATTGTTTTTGC
Seq A exon
GTATTTAACAGAATTTGTAAACCTAGGCAATGTTTCAGCTCTTCGAACTTTCAGAGTCTTGAGAGCTTTGAAAACTATTTCTGTAATCCCAG
Seq C2 exon
GACTAAAGACCATCGTGGGGGCCCTGATCCAGTCAGTGAAGAAGCTCTCTGACGTCATGATCCTCACTGTGTTCTGTCTCAGTGTGTTTGCACTAATTGGACTACAGCTGTTTATGGGCAACTTGAAGCATAAATGTTTCAGGAAGGAACTCGAAGAGAATGAAACATTAGAAAGTATCATGAATACTGCTGAGAGTGAAGAAGAATTGAAAA
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000006639-'6-7,'6-5,7-7
Average complexity
C2
Mappability confidence:
100%=100=80%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
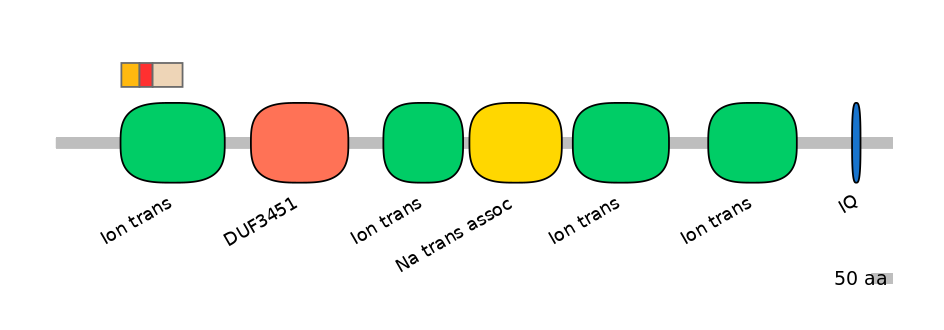
Domain overlap (PFAM):
C1:
PF0052026=Ion_trans=FE(17.3=100)
A:
PF0052026=Ion_trans=FE(12.5=100)
C2:
PF0052026=Ion_trans=FE(28.6=100)
Main Skipping Isoform:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Chicken
(galGal3)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTTCCTCCGTGACCCTTGGAA
R:
GCCCATAAACAGCTGTAGTCCA
Band lengths:
170-262
Functional annotations
There are 1 annotated functions for this event
PMID: 22911851
[MUTEX]. In conditions where the intrinsic properties of the Na(V)1.7 splice variants were similar when expressed alone, co-expression of beta1 subunits had different effects on channel availability that were determined by splicing at either site in the alpha subunit. The identity of exon 5 determined the degree to which beta1 subunits altered voltage-dependence of activation (P_=_0.027). The results could have a significant impact on channel availability. Splicing can change the way that Na(V) channels interact with beta subunits. Because splicing is conserved, its unexpected role in regulating the functional impact of beta subunits may apply to multiple voltage-gated sodium channels, and the full repertoire of beta subunit function may depend on splicing in alpha subunits.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]