DmeEX6020338 @ dm6
Exon Skipping
Gene
FBgn0004237 | Hrb87F
Description
The gene Heterogeneous nuclear ribonucleoprotein at 87F is referred to in FlyBase by the symbol DmelHrb87F (CG12749, FBgn0004237). It is a protein_coding_gene from Dmel. It has 5 annotated transcripts and 5 polypeptides (1 unique). Gene sequence location is 3R:13656846..13660531. Its molecular function is described by 6 unique terms, many of which group under: heterocyclic compound binding; binding; nucleic acid binding; RNA binding; organic cyclic compound binding. It is involved in the biological process described with 10 unique terms, many of which group under: reproductive process; response to stress; multicellular organism reproduction; response to stimulus; multi-organism reproductive process. 21 alleles are reported. The phenotypes of these alleles manifest in: larval neuroblast; ganglion mother cell; eye. The phenotypic classes of alleles include: locomotor behavior defective; lethal - all die before end of larval stage; visible; neuroanatomy defective; lethal; viable. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of very high expression to a trough of moderately high expression. Peak expression observed within 00-18 hour embryonic stages, during late larval stages, during early pupal stages, in adult female stages.
Coordinates
chr3R:13656846-13659648:-
Coord C1 exon
chr3R:13658637-13659648
Coord A exon
chr3R:13658462-13658564
Coord C2 exon
chr3R:13656846-13657125
Length
103 bp
Sequences
Splice sites
3' ss Seq
CATGCATTTATTTTTTACAGGTA
3' ss Score
10.04
5' ss Seq
AAGGTAAAT
5' ss Score
8.88
Exon sequences
Seq C1 exon
GAAGAAGGCTATTGCCAAGCAGGATATGGATCGACAGGGCGGAGGTGGCGGACGCGGAGGTCCTCGAGCTGGCGGTCGCGGTGGTCAGGGTGACCGCGGCCAGGGAGGCGGTGGCTGGGGAGGCCAGAACAGACAGAACGGTGGGGGCAACTGGGGCGGAGCTGGCGGCGGCGGAGGATTCGGCAACAGCGGCGGTAACTTTGGAGGCGGTCAGGGCGGCGGCTCTGGCGGTTGGAATCAGCAAGGCGGAAGCGGAGGTGGTCCATGGAATAACCAGGGTGGCGGCAACGGCGGCTGGAACGGTGGTGGTGGCGGCGGCTACGGCGGCGGAAACAGCAATGGCAGCTGGGGCGGTAACGGTGGTGGAGGTGGTGGTGGCGGTGGCTTCGGAAATGAATACCAGCAGAGCTACGGCGGCGGTCCACAGCGCAACAGCAACTTTGGCAACAACCGTCCAGCTCCTTACAGTCAAGGAGGTGGTGGTGGAGGATTCAACAAGG
Seq A exon
GTAACCAGGGTGGAGGTCAAGGCTTTGCTGGCAACAACTACAACACCGGAGGTGGTGGCCAGGGTGGAAATATGGGAGGCGGCAATAGACGTTACTAGACAAG
Seq C2 exon
ACTGGCCGGGCTACGGACGGAAGCTCCCTCCCGGGTCCGTATACGAAATCTGGGCTGCTCCACGCACCGGTTCGTTTCCACCAGGTATTAGTTAATGGTTTTAGACCGCAAAGCAAGCGGTGGCCCAGGAAGAACTCACTGGTGGACTTTGGTTGGTGGCTCCCTTCTGGCTAACTTGGTGGGTACAGCGGAGGTAAGTGAAGGTAACTAGTCGGTATTCTACTAACACCTCATTTGTTTCTGAATTTTTCAATAAAGTACAACTTTTACTAGCAAGAAC
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0004237-'3-4,'3-1,4-4=AN
Average complexity
A_C3
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref, Alt. Stop)
No structure available
Features
Disorder rate (Iupred):
C1=0.598 A=1.000 C2=NA
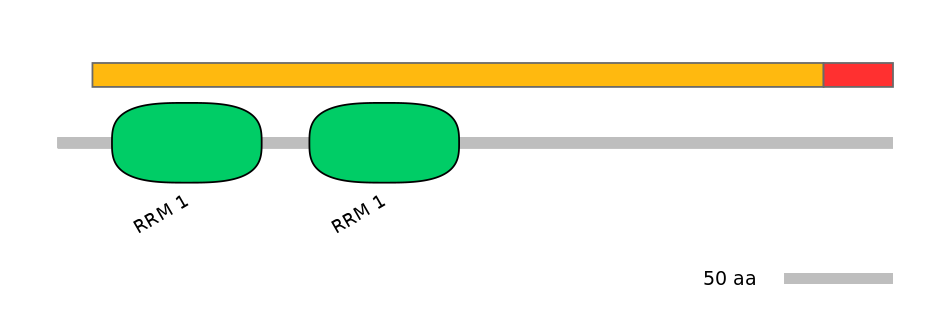
Domain overlap (PFAM):
C1:
PF0007617=RRM_1=WD(100=20.7),PF0007617=RRM_1=WD(100=20.7)
A:
NO
C2:
NA
Main Inclusion Isoform:
FBpp0082316
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0289817, FBpp0302741, FBpp0306562
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Cow
(bosTau6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AACCGTCCAGCTCCTTACAGT
R:
ACCTTCACTTACCTCCGCTGT
Band lengths:
257-360
Functional annotations
There are 1 annotated functions for this event
PMID: 20406423
The inclusion of exon 2 (HsaEX0030279) and exon 9 (HsaEX0030282) generates a predominantly nuclear form, skipping of the exons generates cytosolic forms. The study shows, using isoform-specific antibodies and isoform-specific green fluorescent protein (GFP)-fusion expression constructs, that A2b is the predominant cytoplasmic isoform in neural cells, suggesting that it may play a key role in mRNA trafficking. The differential subcellular distribution patterns of the individual isoforms are determined by the presence or absence of alternative exons that also affect their dynamic behavior in different cellular compartments, as measured by fluorescence correlation spectroscopy. Expression of A2b is also differentially regulated with age, species and cellular development. Furthermore, coinjection of isoform-specific antibodies and labeled RNA into live oligodendrocytes shows that the assembly of RNA granules is impaired by blockade of A2b function. These findings suggest that neural cells modulate mRNA trafficking by regulating alternative splicing of hnRNP A2/B1 and controlling expression levels of A2b, which may be the predominant mediator of cytoplasmic-trafficking functions. B1: inclusion of both. A2: skipping of ex 2. B1b: skipping of ex 9. A2b: skipping of both. Not well in VTS in hg38 nor mm10.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)