DmeEX6023535 @ dm6
Exon Skipping
Gene
FBgn0004903 | Rb97D
Description
The gene Ribonuclear protein at 97D is referred to in FlyBase by the symbol DmelRb97D (CG6354, FBgn0004903). It is a protein_coding_gene from Dmel. It has 10 annotated transcripts and 10 polypeptides (3 unique). Gene sequence location is 3R:26872129..26875742. Its molecular function is described by: RNA binding; mRNA binding. It is involved in the biological process described with: regulation of RNA metabolic process; germ cell development. 14 alleles are reported. The phenotype of these alleles manifest in: axoneme. The phenotypic classes of alleles include: male sterile; lethal. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of moderately high expression to a trough of moderate expression. Peak expression observed within 00-12 hour embryonic stages, during late larval stages, at stages throughout the pupal period, in adult male stages.
Coordinates
chr3R:26872583-26874790:-
Coord C1 exon
chr3R:26874071-26874790
Coord A exon
chr3R:26873860-26873998
Coord C2 exon
chr3R:26872583-26873555
Length
139 bp
Sequences
Splice sites
3' ss Seq
AGACACTCCTCAACCACCAGCAT
3' ss Score
1.29
5' ss Seq
TGGGTGAGT
5' ss Score
8.73
Exon sequences
Seq C1 exon
CCCCTACCAACAACAGCCACCACCGGCGCCTATGTCTGCTCCACCACCAAACTTCAACTACTGGGGACCACCTCCGCCGGCTATGCCGCCCTACTACCAGCAGCCACCGCCGCAGCAGATGAACGGCTGGGGTCCATATCCGCCGCCGCAACAAAATGGCTGGAATGCACCACCACCGCCACCACCGGGTGCCCAGCAGTGGCACGCCAACCAGTGGGGTTGTCCTCCACCAGTGCAACAGGTGCCGCCGGTTGGAGCAGTGCCACCACCCATGGGTCACAATGGACCACCGCCGACGGCTCCAGGTAACTGGAACATGCCGCCCCCAGTGCCGGGTGCAGCTCCTCCATCCCACCAGCAGCAGTCTTCGCAACAGCCAACACCACAACCAAATTTCGGCACTGGCTATCAGCAAAACTACGGAGGCGGACCATCCAAGCACAACAATATGAATGCAAACCGCATGAATCCTTACTCGGCAGGTCCGCCCAACAGCTACC
Seq A exon
CATATACGGGCTACAACGCTGGACCACTGCCGCCTAATGGAAGCGTTCCGCCGCCAACTGGGGCCAAGGCTGTTGGCGTATCCAATGGCTCCGTGGCCACTGGAGGCAGTGCCAACAGCAAATATCGCCGCTAGATTGG
Seq C2 exon
GACAACTAACTACGATTTTACTGTACACTTAAGCTATACCATTGATTACCCTACGATTGACCTAAGCTATGGCAGACTATATATAACTATTTTATACAACGTATTGCGAGGAGCGAGCACTTCTCCAGTTCGAATTACTCTAGACATAATTATGAAACTCTGTATCGTAGAAGAATGCCCAAAATACGATACGAAATTACAACAGGCTCTCTATTTAAATCTAAACCTCTCTCTCTATCTGTAAAACAAGAAGAAGGATTGTGCAATGGTGGAGGATGGAAGAACAATTGCCAATCAAGATCAATTAATTAGACATAAATCACGGAGCTGTAGCCTGAGCTTATTGGCCCAAAACAAGTAGTTTAATCGTAGATGCCTATGTATTACCTAAACCATTCAAATAAGAAGATACGTCCAAACAAAAAAAACACAAGAAACAAGAAACAAGAATATAAGAATCCGGAGCGAAGTCGGATCCCAAATCAAATCAAATCATACGC
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0004903-'4-7,'4-5,6-7=AN
Average complexity
A_C3
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref, Alt. Stop)
No structure available
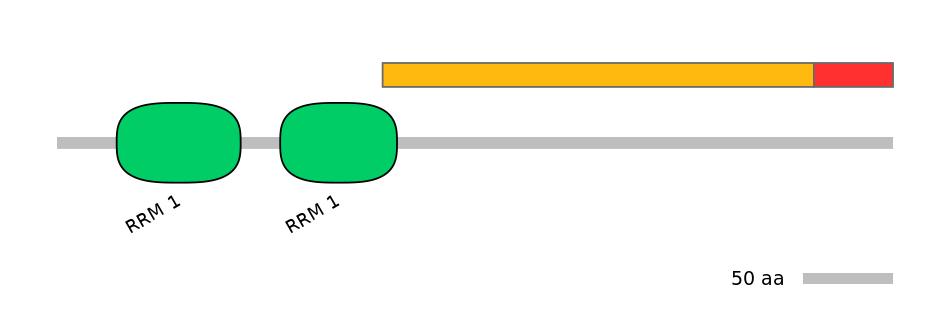
Features
Disorder rate (Iupred):
C1=0.921 A=0.955 C2=1.000
Domain overlap (PFAM):
C1:
PF0007617=RRM_1=PD(12.1=3.3)
A:
NO
C2:
NO
Main Inclusion Isoform:
FBpp0084462
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0084463, FBpp0099739, FBpp0099740, FBpp0099741, FBpp0099742, FBpp0099743, FBpp0099744, FBpp0099745
Other Skipping Isoforms:
NA
Associated events
Conservation
Human
(hg38)
No conservation detected
Cow
(bosTau6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CAACCAAATTTCGGCACTGGC
R:
GAACTGGAGAAGTGCTCGCTC
Band lengths:
246-385
Functional annotations
There are 1 annotated functions for this event
PMID: 20406423
The inclusion of exon 2 (HsaEX0030279) and exon 9 (HsaEX0030282) generates a predominantly nuclear form, skipping of the exons generates cytosolic forms. The study shows, using isoform-specific antibodies and isoform-specific green fluorescent protein (GFP)-fusion expression constructs, that A2b is the predominant cytoplasmic isoform in neural cells, suggesting that it may play a key role in mRNA trafficking. The differential subcellular distribution patterns of the individual isoforms are determined by the presence or absence of alternative exons that also affect their dynamic behavior in different cellular compartments, as measured by fluorescence correlation spectroscopy. Expression of A2b is also differentially regulated with age, species and cellular development. Furthermore, coinjection of isoform-specific antibodies and labeled RNA into live oligodendrocytes shows that the assembly of RNA granules is impaired by blockade of A2b function. These findings suggest that neural cells modulate mRNA trafficking by regulating alternative splicing of hnRNP A2/B1 and controlling expression levels of A2b, which may be the predominant mediator of cytoplasmic-trafficking functions. B1: inclusion of both. A2: skipping of ex 2. B1b: skipping of ex 9. A2b: skipping of both. Not well in VTS in hg38 nor mm10.
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)