BtaEX6032174 @ bosTau6
Exon Skipping
Gene
ENSBTAG00000014947 | PTPN13
Description
Bos taurus protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) (PTPN13), mRNA. [Source:RefSeq mRNA;Acc:NM_174590]
Coordinates
chr6:103498256-103500218:+
Coord C1 exon
chr6:103498256-103498409
Coord A exon
chr6:103499800-103500014
Coord C2 exon
chr6:103500120-103500218
Length
215 bp
Sequences
Splice sites
3' ss Seq
TTTGATACCTTTTTTTCCAGAGA
3' ss Score
6.96
5' ss Seq
CAGGTGAGC
5' ss Score
9.6
Exon sequences
Seq C1 exon
GTGGTTCATCTGTTGTTAGAAAAAGGACAGTCTCCAGCATCCAAAGAACATGTCCTCCAGTGTACGGTCTCTGATCCAGACACCCAAGGTCAAGCCCCAGAAAAAATGATGACGAAAATGACTCATGTCAAAGACTACAGCTTTGTTACCGAAG
Seq A exon
AGAATACATTTGAGGTGAAATTGCTGAAAAATAGCTCAGGCCTAGGATTCAGTTTTTCTCGAGAGGATAATGTTATACCCGAGCAAATGAACACCAGCATAGTAAGGGTTAAAAAGCTCTTTCCTGGACAGCCGGCTGCAGAAAGTGGACAAATTGATGTGGGAGATGTTATCTTAAAAGTGAATGGAGCCTCTTTGAAAGGATTGTCTCAACAG
Seq C2 exon
GAGGTCATATCCGCTCTCAGGGGAACGTCTCCAGAAGTATCCTTGCTCCTCTGTAGACCTCCACCTGGGGTGCTACCAGAAATTGACCCTGCTCTTTCG
VastDB Features
Vast-tools module Information
Secondary ID
ENSBTAG00000014947-'32-38,'32-37,33-38=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.558 A=0.194 C2=0.667
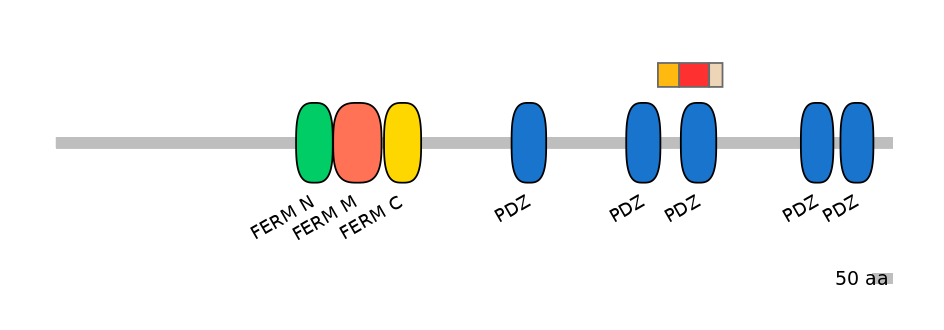
Domain overlap (PFAM):
C1:
PF0059519=PDZ=PD(7.2=11.5)
A:
PF0059519=PDZ=PU(77.9=93.1)
C2:
PF0059519=PDZ=PD(19.8=51.5)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TGGTTCATCTGTTGTTAGAAAAAGGA
R:
CGAAAGAGCAGGGTCAATTTCTG
Band lengths:
252-467
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]