GgaEX1016978 @ galGal4
Exon Skipping
Gene
ENSGALG00000005811 | COL7A1
Description
collagen, type VII, alpha 1 [Source:HGNC Symbol;Acc:HGNC:2214]
Coordinates
chr12:9076923-9081360:+
Coord C1 exon
chr12:9076923-9076958
Coord A exon
chr12:9077087-9077122
Coord C2 exon
chr12:9081319-9081360
Length
36 bp
Sequences
Splice sites
3' ss Seq
ACTGCTTTGCTGTATTTCAGGGG
3' ss Score
10.07
5' ss Seq
CCGGTGAGT
5' ss Score
10.9
Exon sequences
Seq C1 exon
GGCTCACCCGGCCTCCGTGGCATCGCTGGACCGAAG
Seq A exon
GGGGACCCAGGGGAGCCAGGACCACGAGGGGAGCCG
Seq C2 exon
GGTGAGCCGGCGCTGCTGGAGGGCGTGCTGCTGGGTGAACCC
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000005811_MULTIEX2-37/78=36-56
Average complexity
C3
Mappability confidence:
96%=100=96%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=1.000 A=1.000 C2=1.000
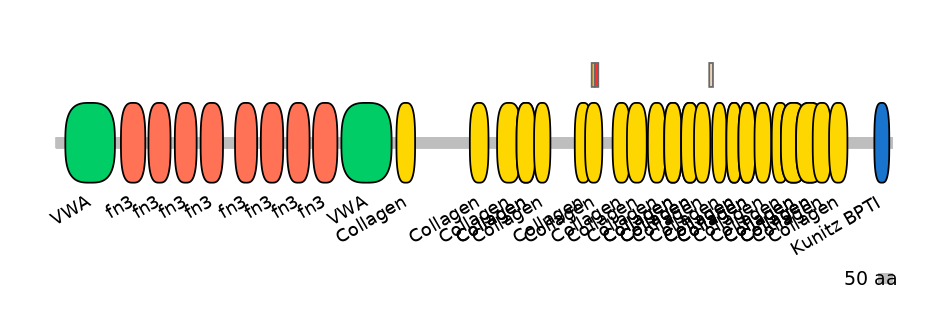
Domain overlap (PFAM):
C1:
PF0139113=Collagen=PD(0.1=0.0),PF0139113=Collagen=FE(18.0=100)
A:
PF0139113=Collagen=FE(18.0=100)
C2:
PF0139113=Collagen=PD(6.9=28.6),PF0139113=Collagen=PU(5.8=21.4)
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
- GgaALTD1006631-1/3
- GgaALTD1006631-2/3
- GgaEX0008179
- GgaEX0008181
- GgaEX1016952
- GgaEX1016953
- GgaEX1016954
- GgaEX1016955
- GgaEX1016956
- GgaEX1016957
- GgaEX1016958
- GgaEX1016959
- GgaEX1016961
- GgaEX1016962
- GgaEX1016963
- GgaEX1016964
- GgaEX1016965
- GgaEX1016966
- GgaEX1016967
- GgaEX1016968
- GgaEX1016969
- GgaEX1016971
- GgaEX1016972
- GgaEX1016973
- GgaEX1016974
- GgaEX1016975
- GgaEX1016976
- GgaEX1016977
- GgaEX1016979
- GgaEX1016981
- GgaEX1016982
- GgaEX1016983
- GgaEX1016985
- GgaEX1016986
- GgaEX1016987
- GgaEX1016988
- GgaEX1016989
- GgaEX1017009
- GgaEX1017010
- GgaINT0022912
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTCCGTGGCATCGCTGGA
R:
GGTTCACCCAGCAGCACG
Band lengths:
65-101
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]