DreEX0065161 @ danRer10
Exon Skipping
Gene
ENSDARG00000011533 | sema6dl
Description
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like [Source:ZFIN;Acc:ZDB-GENE-050417-171]
Coordinates
chr18:40390889-40394121:+
Coord C1 exon
chr18:40390889-40390947
Coord A exon
chr18:40391808-40391963
Coord C2 exon
chr18:40392833-40394121
Length
156 bp
Sequences
Splice sites
3' ss Seq
CAATTTGTTTGTCGCTACAGACA
3' ss Score
6.59
5' ss Seq
AGGGTAAGG
5' ss Score
9.16
Exon sequences
Seq C1 exon
AAGTGGTTATGAGCAAGATGTTGAATATGGCAACACTGCTCACCTCGGGGATTGTCACG
Seq A exon
ACATGGAGTTGCCATCTTCATCAGTCACAGTGCCCGCGACTTCTGAACCCATACAATCACCCCAAACCAACCCCACTCAGACACCACAACTCTTTGAACCCCATGGTGACCCAGCCACTTCACATTCTGTTGAGCATATACCAGGTGTGCTGGAGG
Seq C2 exon
GTGTTTGGGAAATACAAGCAGGTGAATCAAACCAGATGGTGCACATGAACATCCTGATCTCCTGTGTGTTAGCTGCGTTTCTGCTTGGTGCATTCATCGCAGGCATGGTTGTTTACTGCTATAGAGATGCGTTTCTTCGCACTCCACGAAAGATCCAAAAAGATGCAGAGTCGGCACAATCCTGCACTGATTCCACTGGCAGCTTTGCCAAGCTTAACGGTCTGTTTGACAGTCCTGTCAAAGAGTACCAGCCTAATTTGGATACTCCCAAACTCTACACCAACCTGTTGAGCAATGGAAGGGAGGTGCCAACCACTGGAGAAACTAAGACTATGCTCCTGAGTGGGCAGCCACCGGAATTAGCAGCACTGCCTACGCCTGAATCCACACCTGTGCTGCAACAGAAGAGCCTCCAGCCCATTAAGAACCAGTGGGAGAGAGCTCATGGAAAGATGAGTGGCTCCCGCAAAGATGCACCACCCAAAAGCCCACAGTACCTT
VastDB Features
Vast-tools module Information
Secondary ID
ENSDARG00000011533_MULTIEX1-2/2=C1-C2
Average complexity
C2*
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
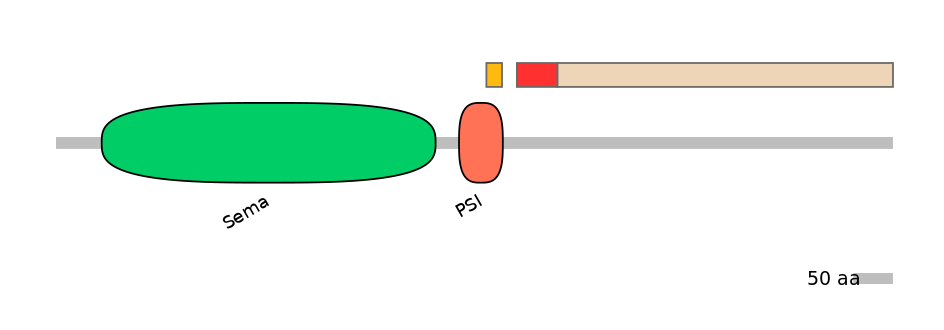
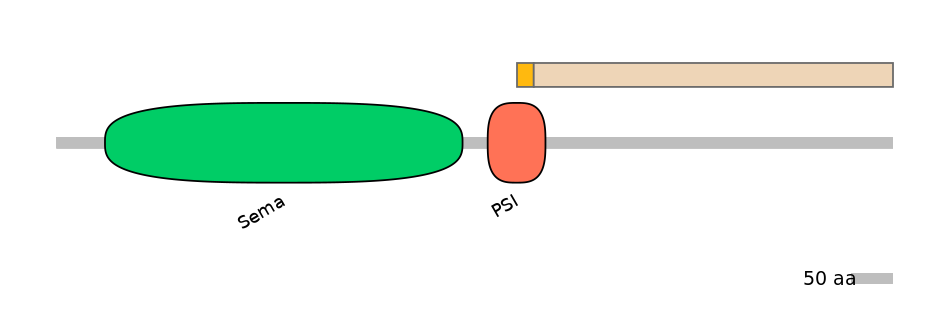
Features
Disorder rate (Iupred):
C1=0.037 A=0.733 C2=0.801
Domain overlap (PFAM):
C1:
PF0143720=PSI=FE(28.6=100)
A:
NO
C2:
PF0143720=PSI=PD(20.0=3.3)
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
CTCACCTCGGGGATTGTCACG
R:
ACAGACCGTTAAGCTTGGCAA
Band lengths:
246-402
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]