DmeEX6002886 @ dm6
Exon Skipping
Gene
FBgn0264255 | para
Description
NA
Coordinates
chrX:16466495-16473827:-
Coord C1 exon
chrX:16473582-16473827
Coord A exon
chrX:16471214-16471484
Coord C2 exon
chrX:16466495-16466799
Length
271 bp
Sequences
Splice sites
3' ss Seq
GACTTCTTTTACCATTCCAGTGG
3' ss Score
7.64
5' ss Seq
TAGGTAAGT
5' ss Score
9.66
Exon sequences
Seq C1 exon
GTGGACAAGCAACCAATTCGTGAAACGAACATCTACATGTATTTATATTTCGTATTCTTCATCATATTTGGATCATTTTTCACACTCAATCTGTTCATTGGTGTTATCATTGATAATTTTAATGAGCAAAAGAAAAAAGCAGGTGGATCATTAGAAATGTTCATGACAGAAGATCAGAAAAAGTACTATAATGCTATGAAAAAGATGGGCTCTAAAAAACCATTAAAAGCCATTCCAAGACCAAGG
Seq A exon
TGGCGACCACAAGCAATAGTCTTTGAAATAGTAACCGATAAGAAATTCGATATAATCATTATGTTATTCATTGGTCTGAACATGTTCACCATGACCCTCGATCGTTACGATGCGTCGGACACGTATAACGCGGTCCTAGACTATCTCAATGCGATATTCGTAGTTATTTTCAGTTCCGAATGTCTATTAAAAATATTCGCTTTACGATATCACTATTTTATTGAGCCATGGAATTTATTTGATGTAGTAGTTGTCATTTTATCCATCTTAG
Seq C2 exon
GTCTTGTACTTAGCGATATTATCGAGAAGTACTTCGTGTCGCCGACCCTGCTCCGAGTGGTGCGTGTGGCGAAAGTGGGCCGTGTCCTTCGACTGGTGAAGGGAGCCAAGGGCATTCGGACACTGCTCTTCGCGTTGGCCATGTCGCTGCCGGCCCTGTTCAACATCTGCCTGCTGCTGTTCCTGGTCATGTTCATCTTTGCCATTTTCGGCATGTCGTTCTTCATGCACGTGAAGGAGAAGAGCGGCATCAACGACGTCTACAACTTCAAGACCTTTGGCCAGAGCATGATCCTGCTCTTTCAG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0264255-'39-44,'39-43,40-44=AN
Average complexity
A_S
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF0052026=Ion_trans=PD(15.7=42.7)
A:
PF0052026=Ion_trans=PU(20.8=47.3)
C2:
PF0052026=Ion_trans=FE(48.8=100)
Main Inclusion Isoform:
FBpp0303597
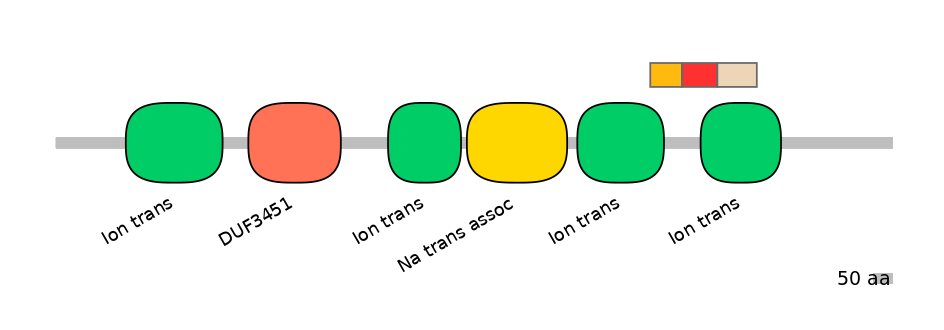
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0074073, FBpp0110321, FBpp0110322, FBpp0288560, FBpp0292672, FBpp0292673, FBpp0292674, FBpp0292675, FBpp0292676, FBpp0292677, FBpp0292678, FBpp0292679, FBpp0292680, FBpp0292681, FBpp0292682, FBpp0292683, FBpp0292684, FBpp0292685, FBpp0292686, FBpp0292687, FBpp0292688, FBpp0292689, FBpp0292690, FBpp0292691, FBpp0292692, FBpp0292693, FBpp0292694, FBpp0292695, FBpp0292696, FBpp0292697, FBpp0292698, FBpp0292699, FBpp0292700, FBpp0292701, FBpp0292702, FBpp0292703, FBpp0292704, FBpp0292705, FBpp0292706, FBpp0292707, FBpp0292708, FBpp0292709, FBpp0292710, FBpp0292712, FBpp0292713, FBpp0292716, FBpp0292717, FBpp0292718, FBpp0292719, FBpp0292720, FBpp0292721, FBpp0292723, FBpp0309614, FBpp0311217
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Cow
(bosTau6)
No conservation detected
Chicken
(galGal3)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
GTGGACAAGCAACCAATTCGTG
R:
GTCGGCGACACGAAGTACTTC
Band lengths:
292-563
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)