DmeEX6014697 @ dm6
Exon Skipping
Gene
FBgn0035533 | Cip4
Description
The gene Cdc42-interacting protein 4 is referred to in FlyBase by the symbol DmelCip4 (CG15015, FBgn0035533). It is a protein_coding_gene from Dmel. It has 12 annotated transcripts and 12 polypeptides (10 unique). Gene sequence location is 3L:4322491..4364102. Its molecular function is described by: GTPase activating protein binding; lipid binding; phospholipid binding; protein binding. It is involved in the biological process described with 13 unique terms, many of which group under: macromolecule localization; plasma membrane organization; membrane organization; mesoderm development; germ cell development. 36 alleles are reported. The phenotypes of these alleles manifest in: multicellular structure; organism; cell junction; cellular anatomical entity; integumentary system. The phenotypic classes of alleles include: increased mortality during development; phenotype; increased mortality; neuroanatomy defective. Summary of modENCODE Temporal Expression Profile: Temporal profile ranges from a peak of high expression to a trough of moderate expression. Peak expression observed within 00-18 hour embryonic stages, during early pupal stages.
Coordinates
chr3L:4325061-4327702:-
Coord C1 exon
chr3L:4327233-4327702
Coord A exon
chr3L:4326984-4327170
Coord C2 exon
chr3L:4325061-4325410
Length
187 bp
Sequences
Splice sites
3' ss Seq
TCTCTATGTCCGCCTTGCAGATA
3' ss Score
11.26
5' ss Seq
AAGGTGAGG
5' ss Score
9.16
Exon sequences
Seq C1 exon
AAATGCCTTAGCGATGGTGCCACCCTGCAGCAGAACCTCACCACACAGCTCTCCTCGCTGGACCGGGCCAAGCGGAACTACGAGAAGGCCTACCGTGACTCGGAGAAGGCGGTGGACAGCTATAAGCGGGCAGACATGGACCTCAATCTCAGCCGGGCCGAGGTGGAGCGCTACAAGAACGTGATGACGTCCAAGATCCAGCAGTCGGACGATGCGAAGAACGAGTACGCTAACCAGCTACAGAAGACGAACAATCTGCAGCAGCAACACTACAGCATGCTGCTGCCCTCGGTCCTCAATCGGCTGCAGGAGCTGGACGAGAAACGCACCCGTGGCTTCAGGGAGTTCATTGTGGGAGCGGCGGATGTGGAGTCATCGGTGGCGCCAATCATAGCCCGCTGCATGGAGGGTATCGTGAAGGCCGGCGAGTCCATCAACGAAAAGGAGGATACCTTCAAAGTCATAGAAAG
Seq A exon
ATATCAATCTGGTTTCACGCCACCAAGGGACATACCCTTCGAGGATCTGTCCAAGTGCGATCCGGATTCCGTGCAGGACTCACACTACAGCAACTCGACATCGAACCACCTGACCATTAGAGGCACGATGAGTGCCAACAAGCTGAAGAAACGCGTGGGCATTTTTAACATATTCGGCAGCAATAAG
Seq C2 exon
AATTCCCTGACTGCGGATGGACAAAAGGAGGACTTCAGCGATCTGCCACCGAATCAACGAAGAAAGAAACTGCAGGCGAAGATCGCCGAACTGACACAGAATATCGCCCAGGAAACAAAAGCACGGGATGGCCTGATGAAGATGAAGATCGTCTATGAGGCGAACTCATCGCTGGGCAATCCCATGACCGTCGAGGGACAACTGAACGAGTCGGAACACAAGTTGGAGAAGCTGAAAGTGGATCTAAAGAAGTACCAGGGCTTCTTGGAGAAGGCAAGCCAAGTGCCGACGGCCACCAGTAGTCCGCAGGCGAGTCGAAATCAATTGCAAAACGGTCACCGAACCTCTAG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0035533-'18-15,'18-11,19-15=AN
Average complexity
A_C1
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.185 A=0.450 C2=0.865
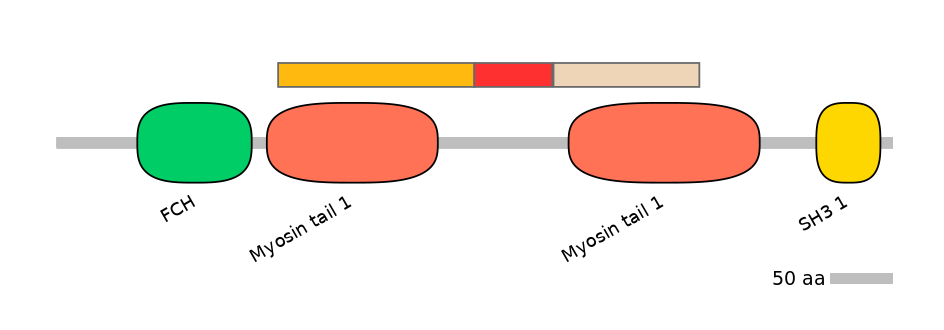
Domain overlap (PFAM):
C1:
PF0157614=Myosin_tail_1=PD(92.7=80.9)
A:
PF154561=Uds1=PU(0.1=0.0)
C2:
PF154561=Uds1=PD(97.7=78.7),PF0157614=Myosin_tail_1=PU(91.4=98.1),PF0218511=HR1=WD(100=64.8)
Main Inclusion Isoform:
FBpp0112051
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0073195, FBpp0297231, FBpp0297232, FBpp0297233, FBpp0297491, FBpp0297492, FBpp0300521, FBpp0300522, FBpp0305747, FBpp0305748
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
CAGGGAGTTCATTGTGGGAGC
R:
TGCTTTTGTTTCCTGGGCGAT
Band lengths:
255-442
Functional annotations
There are 3 annotated functions for this event
PMID: 22749400
Differential PPI with CDC42 and TC10. The F-BAR protein CIP-4 has an important role in endocytosis. The second coiled-coil domain of CIP4 (residues 332?425) is required for interaction with the GTPases CDC-42 and TC-10. [From Buljan et al, Mol Cell 2012, Table 2]
PMID: 12242347
Inclusion of exon 10 (MmuEX0048979) makes it interact with TC10 in vitro and in vivo.
PMID: 27851958
The splicing switch in 4 events (CLTC, TMED2, TRIP10, SNAP23) results in multiple structural and functional defects, including transverse tubule (T-tubule) disruption and dihydropyridine receptor alpha (DHPR) and Ryr1 mislocalization, impairing excitation-contraction coupling, calcium handling, and force generation
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)