GgaEX0017948 @ galGal4
Exon Skipping
Gene
ENSGALG00000011963 | PRDM5
Description
PR domain containing 5 [Source:HGNC Symbol;Acc:HGNC:9349]
Coordinates
chr4:53686928-53695187:+
Coord C1 exon
chr4:53686928-53687012
Coord A exon
chr4:53688295-53688452
Coord C2 exon
chr4:53695094-53695187
Length
158 bp
Sequences
Splice sites
3' ss Seq
CCCCCACTCATTTTCTGCAGAAA
3' ss Score
10.02
5' ss Seq
AAGGTAAAG
5' ss Score
9.06
Exon sequences
Seq C1 exon
GTTCATGAGACCTTTGAGTGTCAGGAATGTGACAAGAGATTTATTTCAGCTAACCAGCTAAAACGCCATATGATAACTCATTCAG
Seq A exon
AAAAACGCCCCTACACCTGCGAGGTTTGCAATAAGTCCTTCAAACGACTTGATCAAGTGACTGCACACAAAATAATACACAGTGAAGATAAACCTTATAAATGCAAGCTTTGTGGAAAGGGATTTGCCCACAGAAATGTTTACAAGAATCACAAGAAG
Seq C2 exon
ACCCATTCTGAAGAGAGACCATTCCAATGTGAGGAATGCAAAGCTTTGTTCCGAACCCCATTCTCTCTACAAAGACATCTACTAATTCATAACA
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000011963_MULTIEX2-1/2=C1-C2
Average complexity
C2
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.080 A=0.000 C2=0.000
Domain overlap (PFAM):
C1:
PF121713=zf-C2H2_jaz=PU(75.8=86.2)
A:
PF134651=zf-H2C2_2=PD(57.7=28.3),PF134651=zf-H2C2_2=WD(100=47.2)
C2:
PF134651=zf-H2C2_2=PD(85.7=56.2),PF134651=zf-H2C2_2=PU(38.5=31.2)
Main Inclusion Isoform:
ENSGALT00000019510fB2109
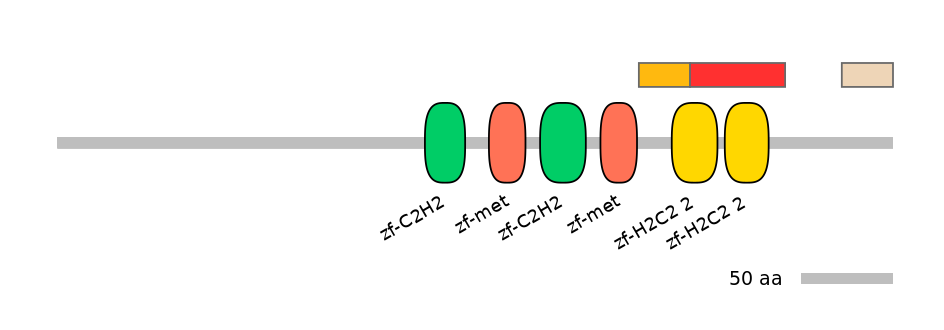
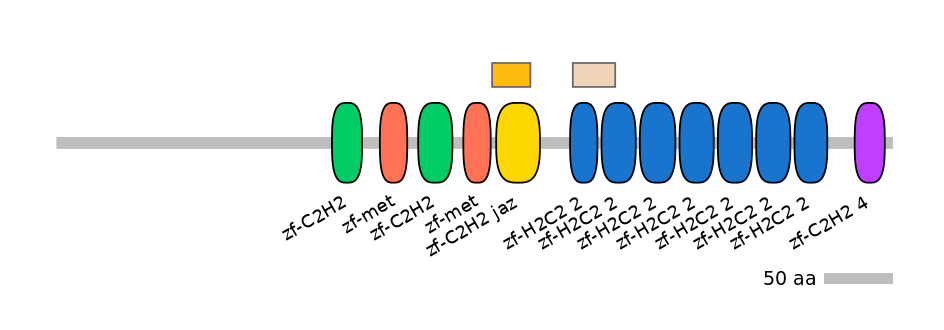
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
TCATGAGACCTTTGAGTGTCAGG
R:
GATGTCTTTGTAGAGAGAATGGGGT
Band lengths:
162-320
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]