RnoEX0070346 @ rn6
Exon Skipping
Gene
ENSRNOG00000033343 | Prss36
Description
serine protease 36 [Source:RGD Symbol;Acc:1593186]
Coordinates
chr1:199380093-199390395:-
Coord C1 exon
chr1:199390229-199390395
Coord A exon
chr1:199382498-199382736
Coord C2 exon
chr1:199380093-199380214
Length
239 bp
Sequences
Splice sites
3' ss Seq
CAACCTCGCACCTCTGACAGCTC
3' ss Score
6.18
5' ss Seq
GGGGTGCGC
5' ss Score
3.75
Exon sequences
Seq C1 exon
ACCCTCTGCCTGTCCCCTGGGTTCTACAGGAAGTAGAGCTAAAGTTGCTGGGAGAGACTGCCTGTCAATGTCTCTACAGTCGGCCTGGCCCTTTCAACCTGACTTTGCAGCTGTTGCCAGGAATGCTGTGTGCTGGCTATCCAGAGGGACGAAGGGACACCTGCCAG
Seq A exon
CTCTCTCCACGATTTCGAGACCTGGCGGGTGCTGCTGCCCTCGCGTCCGGAGGAGAAGAGGGTCGTGCGCCTCGTGGCGCACGAGAACGCTTCCAGGAACTTCGCTTCAGACCTGGCGCTACTGCAGCTGAGGACGCGCGTGAACCTGACTGCGGCTCCGAGTGCTGTGTGCCTGCCCCACCGCGAACACTATTTTCTGCCCGGGAGCCGCTGCCGCCTAGCTCGCTGGGGACACGGGG
Seq C2 exon
TTCCTGTAGCTGCTGCTGTCTCCATCTTGACACCACGACTCTGTCACTGCCTCTATCAGGGCGCTCTGACTCCTGGAACCTTCTGTGTCTTCTATACTGAAGAGCAAGAGGATAGATGTGAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSRNOG00000033343_MULTIEX1-4/9=C1-9
Average complexity
C3
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.012 C2=0.000
Domain overlap (PFAM):
C1:
PF0008921=Trypsin=FE(22.9=100)
A:
PF0008921=Trypsin=FE(36.2=100)
C2:
PF0008921=Trypsin=FE(20.1=100)
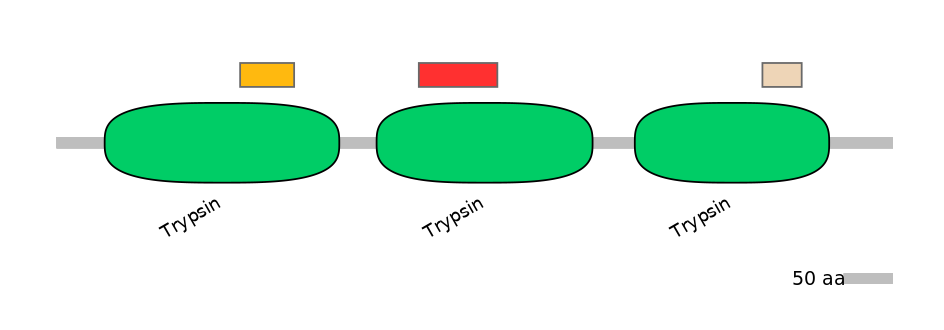
Main Skipping Isoform:
ENSRNOT00000026587fB11469
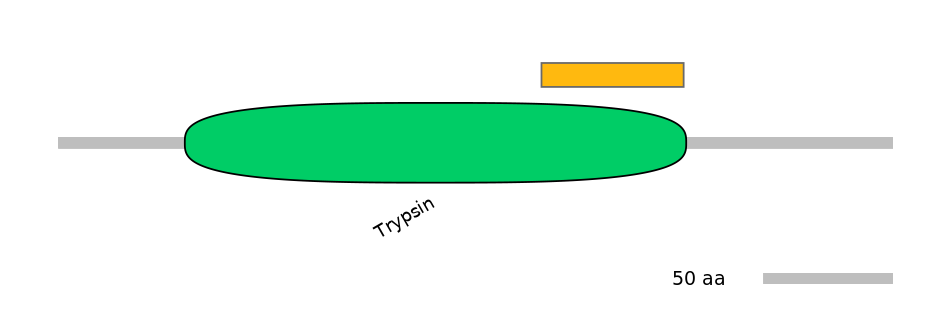
Other Skipping Isoforms:
NA
Associated events
Conservation
Mouse
(mm9)
No conservation detected
Chicken
(galGal4)
No conservation detected
Chicken
(galGal3)
No conservation detected
Zebrafish
(danRer10)
No conservation detected
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
CTCTGCCTGTCCCCTGGG
R:
CACATCTATCCTCTTGCTCTTCAGT
Band lengths:
284-523
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]