DmeEX0008584 @ dm6
Exon Skipping
Gene
FBgn0265487 | mbl
Description
The gene muscleblind is referred to in FlyBase by the symbol Dmelmbl (CG33197, FBgn0265487). It is a protein_coding_gene from Dmel. It has 19 annotated transcripts and 19 polypeptides (18 unique). Gene sequence location is 2R:17216549..17379376. Its molecular function is described by: RNA binding; metal ion binding; nucleic acid binding. It is involved in the biological process described with 9 unique terms, many of which group under: regulation of nucleobase-containing compound metabolic process; reproductive process; programmed cell death; apoptotic process; organic cyclic compound metabolic process. 93 alleles are reported. The phenotypes of these alleles manifest in: fascicle; larva; rhabdomere; visceral muscle; cytoskeleton. The phenotypic classes of alleles include: increased mortality during development; some die during embryonic stage; phenotype; courtship behavior defective.
Coordinates
chr2R:17275410-17352305:+
Coord C1 exon
chr2R:17275410-17276063
Coord A exon
chr2R:17351094-17351264
Coord C2 exon
chr2R:17352129-17352305
Length
171 bp
Sequences
Splice sites
3' ss Seq
TCGCTTGAATTTACTTGCAGGGC
3' ss Score
7.39
5' ss Seq
GTGGTAAGT
5' ss Score
10.36
Exon sequences
Seq C1 exon
CCACAAGCTTTGTACTCCACAAAAACGCTAACGAGCAAAATCGATAAAGACCAGATAGCAAATGAAATCATACCATCTGCCATAGATCTATGCTATAAACATATATAGGTGCTGATCTGTGCTCTTGAATTTAGTGTTTTTTGTGTGTGCGTGTCGCGAGAGGAGCACTCAAAACCAAAAAAAAAAAAAAAAACGTATAGTAAAACAAAAAACTATCTCTAGTCGCTAGGCTATAACTATACAAACTCCAGCTATCCCAGATCCAGATCCGTAAACAGCAAATAGTTATATATCTATAACCCCAAAATACGATGGCCAACGTTGTCAATATGAACAGCCTGCTCAACGGCAAGGATTCGCGCTGGCTGCAATTGGAGGTCTGTCGCGAGTTCCAGCGCAACAAATGCTCGCGCCAGGACACCGAATGCAAGTTCGCCCATCCCCCGGCCAACGTGGAGGTCCAGAACGGCAAGGTCACCGCCTGCTACGACAGCATCAAG
Seq A exon
GGCCGCTGTAATCGTGATAAACCGCCGTGCAAATACTTTCATCCACCACAGCATCTGAAGGATCAGTTATTGATAAATGGACGCAATCATTTGGCCCTCAAGAATGCACTGATGCAACAGATGGGCATCGCGCCCGGCCAACCGGTCATATCGGGCCAAGTGCCAGCCGTG
Seq C2 exon
GCAACAAACCCCTATCTGACCGGCATTCCGGCCAACTCGTACAGTCCGTACTACACAACGGGACACCTGGTGCCCGCCTTGCTGGGTCCGGATCCCGTGACCTCCCAACTGGGACCCGTGGTGCCCCAAACAGTGCAGGTGGCACAGCAGAAAATACCACGTTCCGACAGATTAGAG
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0265487-'10-19,'10-18,17-19
Average complexity
C1
Mappability confidence:
100%=100=100%
Protein Impact
Alternative protein isoforms (Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.032 A=0.112 C2=0.299
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
PF0064219=zf-CCCH=PU(11.1=5.1)
Main Inclusion Isoform:
FBpp0307871
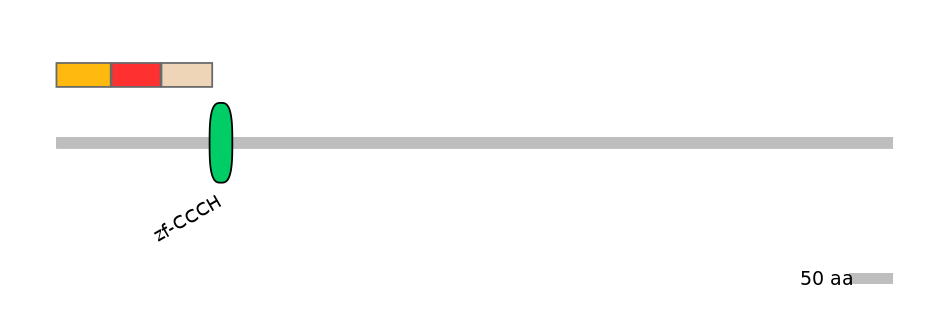
Main Skipping Isoform:
NA
Other Inclusion Isoforms:
FBpp0086143, FBpp0086144, FBpp0086146, FBpp0297556, FBpp0297558, FBpp0297559, FBpp0297561, FBpp0301759, FBpp0301760, FBpp0302029, FBpp0307872, FBpp0307873
Other Skipping Isoforms:
NA
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
ACGATGGCCAACGTTGTCAAT
R:
GTACGGACTGTACGAGTTGGC
Band lengths:
243-414
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)