GgaEX0011327 @ galGal4
Exon Skipping
Gene
ENSGALG00000007943 | lasp-2
Description
nebulette non-muscle isoform [Source:RefSeq peptide;Acc:NP_989819]
Coordinates
chr2:18204652-18225701:+
Coord C1 exon
chr2:18204652-18204762
Coord A exon
chr2:18207593-18207703
Coord C2 exon
chr2:18225530-18225701
Length
111 bp
Sequences
Splice sites
3' ss Seq
ATCTCTCTTTAAATCCTTAGAAA
3' ss Score
7.1
5' ss Seq
GAGGTACGT
5' ss Score
10.2
Exon sequences
Seq C1 exon
AAAATATATCGCAAAGATTTTGAAGAAAGTCTTAAGGGAAGGCCCTCACTGGATTTAGACAAAACCCCAGAGTTCTTGCATATAAAGCAAGTCACTAATCTTCTGAAGGAG
Seq A exon
AAAGAGTATAGAAAAGATTTGGAAGAATGGATGAAAGGAAAAGGAATGACAGTGTTTGAAGATACGCCAGATTTGATTAGAGTTAAAAATGCGGCTCAGATACTAAATGAG
Seq C2 exon
GTTAAATATCATGAAGATTTTGAGAAGACAAAGGGCAGGGGTTTCACGCCAGTGGTGGATGATCCTATAACAGAGCGGGTGCGGAAAAACACCCAAGTTGTAAGTGATGCAGCATATAAAGGGGTGCACCCACATATAGTTGAAATGGATAGAAGACCTGGAATAATTGTTG
VastDB Features
Vast-tools module Information
Secondary ID
ENSGALG00000007943_MULTIEX1-17/26=16-C2
Average complexity
C3
Mappability confidence:
90%=100=91%
Protein Impact
Alternative protein isoforms (No Ref)
No structure available
Features
Disorder rate (Iupred):
C1=0.054 A=0.000 C2=0.428
Domain overlap (PFAM):
C1:
PF0088013=Nebulin=PD(37.9=29.7),PF0088013=Nebulin=PU(55.2=43.2)
A:
PF0088013=Nebulin=PD(37.9=29.7),PF0088013=Nebulin=PU(55.2=43.2)
C2:
PF0088013=Nebulin=PD(37.9=19.0),PF0088013=Nebulin=WD(100=41.4)
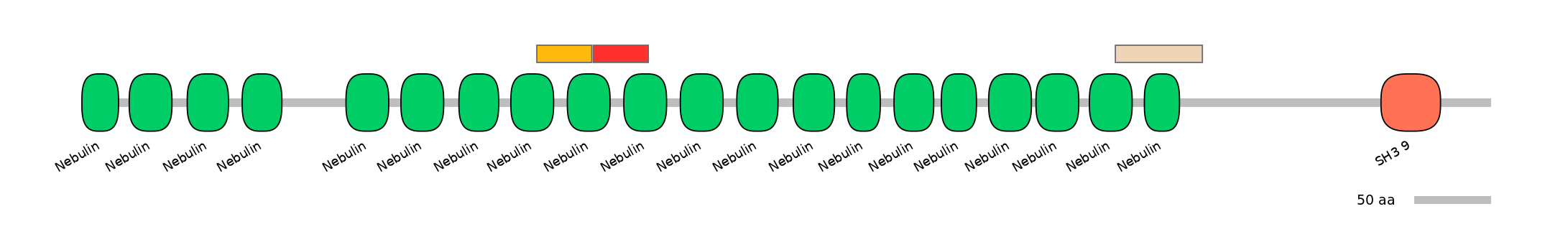
Main Skipping Isoform:
ENSGALT00000012901fB1254
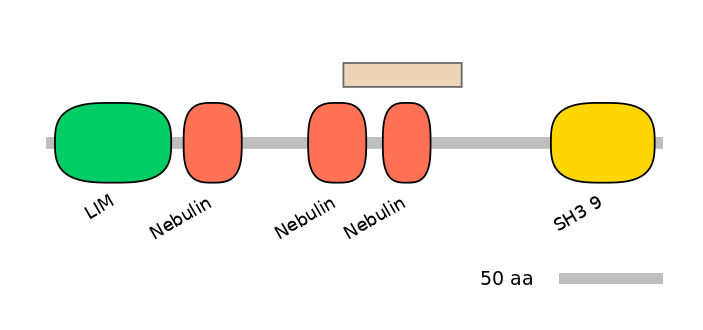
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
- GgaALTD0003113-3/4
- GgaALTD0003113-4/4
- GgaEX0011325
- GgaEX0011326
- GgaEX0011328
- GgaEX0011330
- GgaEX0011331
- GgaEX0028345
- GgaEX0028346
- GgaEX0028347
- GgaEX0028348
- GgaEX0028349
- GgaEX0028350
- GgaEX0028351
- GgaEX0028352
- GgaEX0028353
- GgaEX0028354
- GgaEX0028355
- GgaEX1066197
- GgaEX1066198
- GgaEX1066199
- GgaEX1066200
- GgaEX7001767
- GgaEX7001768
- GgaINT1030833
Other assemblies
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
AAGAAAGTCTTAAGGGAAGGCCC
R:
TCCAGGTCTTCTATCCATTTCAACT
Band lengths:
251-362
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]