DmeEX6009751 @ dm6
Exon Skipping
Gene
FBgn0261698 | CG42732
Description
The gene slowpoke 2 is referred to in FlyBase by the symbol DmelSLO2 (CG42732, FBgn0261698). It is a protein_coding_gene from Dmel. It has 10 annotated transcripts and 10 polypeptides (9 unique). Gene sequence location is 2R:10322195..10416996. Its molecular function is described by: calcium-activated potassium channel activity; outward rectifier potassium channel activity; intracellular sodium activated potassium channel activity. It is involved in the biological process described with: potassium ion transmembrane transport; potassium ion transport. 21 alleles are reported. The phenotypes of these alleles manifest in: mesothoracic tergum; ganglion mother cell; trichogen cell. The phenotypic classes of alleles include: phenotype; increased mortality; increased mortality during development; some die during P-stage.
Coordinates
chr2R:10340123-10412896:-
Coord C1 exon
chr2R:10412236-10412896
Coord A exon
chr2R:10357515-10357708
Coord C2 exon
chr2R:10340123-10340202
Length
194 bp
Sequences
Splice sites
3' ss Seq
TTCGATTGCATAACCTGCAGCGC
3' ss Score
4.96
5' ss Seq
CAGGTGAGT
5' ss Score
10.67
Exon sequences
Seq C1 exon
GGTAAGGGGCATTGTTCGACCAGAAGGACACTCGCCAATCTGTGGACGCGCCTACGCTCGCCGCGGCCCAATCGGGCCACTGGCGGCTATCTGCTGACGGGCACAAGTGCGCCGCGTGACGATGAGTTCGTCGCAGGACGGAGGCATCTTCTGGATCAGGGCAAGGATAAGGATAAGGAGCAGCAGCAGCTGGGGCGGCGGGCATACGCACCTGCCGATGAGTGCTTAATTAATGTGACGGACTTTAGTGATATACAAATAAGTGAGGGGACTGGCGAGGCAGTGGCCAAGACCATCACCGAGCTGAGAGTGCCCACCATAACGACCACAAACAACAGCACCACCATCCACAGCAACAGCAACAGCAACAGCAACAGCAACAGCAACAGCAGCAACATCAACAGCAACAACAATAGCAACATGTCGCGCCTTATACAACCGCGATATCGTTTTCGGGATCTATTACTGGGCGATTTCTCATTTAACGACGACGGCGAAAG
Seq A exon
CGCGCCGAGGAGAACAAGCGCAAGAAGCGCGTGGAGCAGCAGCGCCAGCAGCAACGCCAGCGGAGGGGGGCCAAGGTGTTCAAGTGTGTCAGTGATTCCACCGGCCACCTGACCTTGGCCCCCTCGCGCCTCTTCGAGCGCACGCCGATGAGCCCCTCGGACAGTCTGGACGAGATCGCTCTCCAGATCCCCAG
Seq C2 exon
AGTTCGGGTGGAGTACTATGTGAACGAAAACACATTCAAAGAAAGACTGCAACTCTATTTCATTAAGAATCAGCGTTCAA
VastDB Features
Vast-tools module Information
Secondary ID
FBgn0261698-'2-15,'2-10,8-15=AN
Average complexity
A_C2
Mappability confidence:
100%=100=100%
Protein Impact
In the CDS, with uncertain impact
No structure available
Features
Disorder rate (Iupred):
C1=0.000 A=0.272 C2=0.000
Domain overlap (PFAM):
C1:
NO
A:
NO
C2:
NO
Main Inclusion Isoform:
FBpp0302970
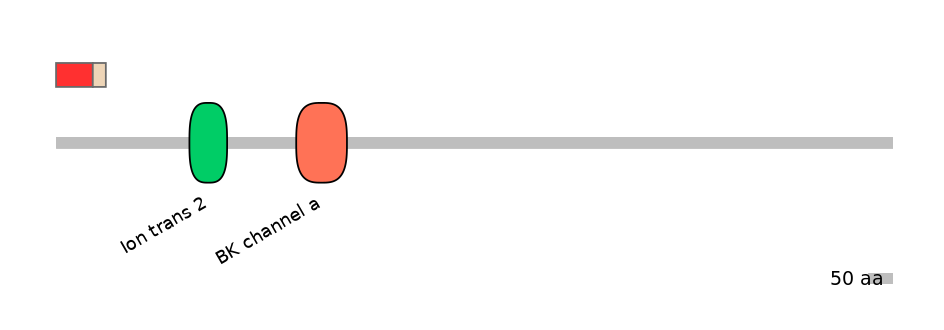
Main Skipping Isoform:
FBpp0302905
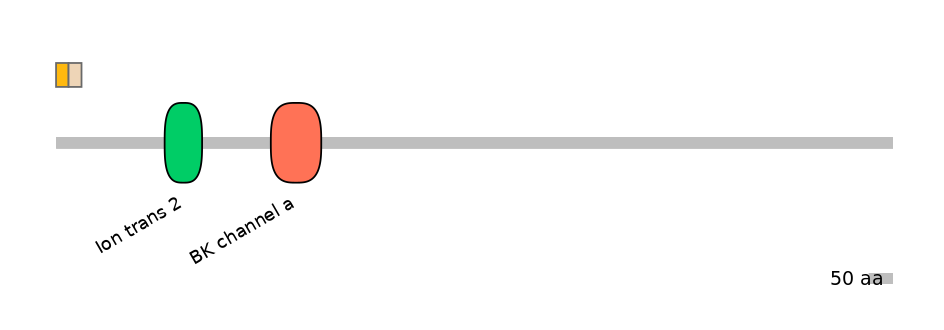
Other Inclusion Isoforms:
FBpp0292201
Other Skipping Isoforms:
FBpp0292202, FBpp0297088, FBpp0303466
Associated events
Conservation
Primers PCR
Suggestions for RT-PCR validation
F:
CAGTGGCCAAGACCATCACC
R:
CGTTCACATAGTACTCCACCCG
Band lengths:
246-440
Functional annotations
There are 0 annotated functions for this event
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Neural diversity
- Neurogenesis
- Neuronal activity
- Splicing factor regulation (brain)
- Splicing factor regulation (SL2)