MmuEX1010315 @ mm10
Exon Skipping
Gene
ENSMUSG00000015095 | Fbxw5
Description
F-box and WD-40 domain protein 5 [Source:MGI Symbol;Acc:MGI:1354731]
Coordinates
chr2:25317464-25505471:+
Coord C1 exon
chr2:25317464-25317485
Coord A exon
chr2:25502871-25503045
Coord C2 exon
chr2:25504710-25505471
Length
175 bp
Sequences
Splice sites
3' ss Seq
CTGCAGCCTTTCTGGAACAGATC
3' ss Score
5.31
5' ss Seq
TAGGTAAGT
5' ss Score
9.66
Exon sequences
Seq C1 exon
ACAACACACACACACACACAAC
Seq A exon
ATCTGGAACAACGACCTGACCATCTCACTGCTGCACAGTGCAGATATGAGGCCATACAACTGGAGTTACACCCAGTTTTCCCAGTTCAACCAGGATGACTCACTGCTGCTGGCCTCAGGGGTGTTCCTGGGGCCACATAACTCCTCCTCAGGCGAGATAGCTGTCATCAGCTTAG
Seq C2 exon
TGGGGCTGAAGATCGGCATGGCTATATCTGGGACCGCCACTACAACATCTGCCTAGCCAAGCTGCGGCATGAGGATGTGGTCAACTCCGTGGCTTTTAGCCCCCAGGAGCAGGAACTCTTGCTGACAGCCAGCGACGATGCCACTATCAAAGCCTGGCGTTCACCACGCATTGTTCGTGTTCTGCAGGCTCCACGCCCTCGCCCCCGCCCTCGCCCCCGCCCCTTCTTCTCCTGGTTTGCCAGCCATAGGCGCTGAAGGCTTCAGTGATTTGAGCCATTGGAACCTACTGGGGAAAATGGTACCCCGTGGTTCTTTATTGCACTGGGGCAGAGTAGCTCATAGCAGCAATGCCTTAGGAGGGGCAGCCTGACCCCAACACACACACACACACACACACACACAAAACCTGCTCACTTAGCCATAAGGCTTAGCACTAGGGTAGCCCTTGTTGGCAGCACCAGATCCCATGCTAGCCTCTTGGCTAGCTTGGGGTACACAG
VastDB Features
Vast-tools module Information
Secondary ID
ENSMUSG00000015095_MULTIEX1-3/8=C1-C2
Average complexity
C3*
Mappability confidence:
100%=100=100%
Protein Impact
ORF disruption upon sequence exclusion
No structure available
Features
Disorder rate (Iupred):
C1=NA A=0.000 C2=0.020
Domain overlap (PFAM):
C1:
NA
A:
PF0040027=WD40=PD(5.1=3.4)
C2:
PF0040027=WD40=WD(100=43.0)
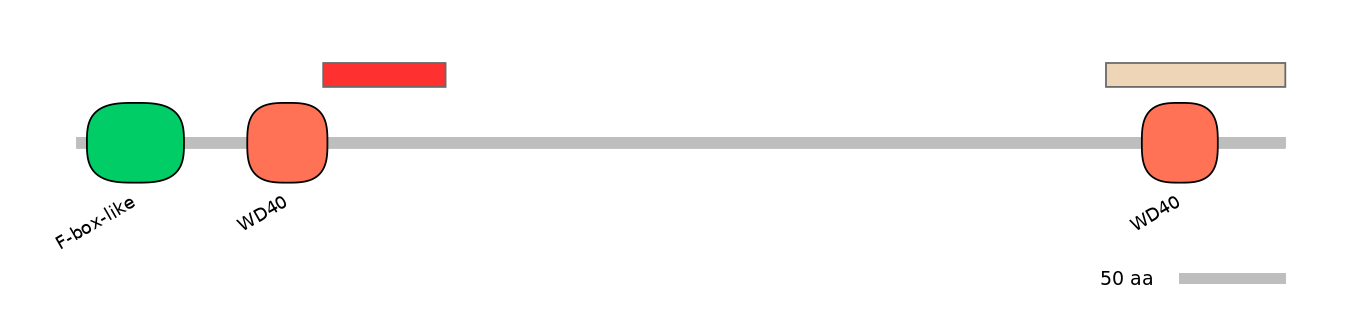
Other Inclusion Isoforms:
NA
Other Skipping Isoforms:
NA
Associated events
- MmuALTA0002556-1/2
- MmuALTA0002556-2/2
- MmuALTA1004254-1/3
- MmuALTA1004254-2/3
- MmuALTA1004256-2/5
- MmuALTA1004256-3/5
- MmuALTD0009496-1/2
- MmuALTD0009496-2/2
- MmuALTD0011247-3/4
- MmuALTD0011247-4/4
- MmuALTD0015071-1/3
- MmuALTD0015071-3/3
- MmuALTD1003597-1/2
- MmuALTD1003597-2/2
- MmuEX0000755
- MmuEX0000756
- MmuEX0002972
- MmuEX0007424
- MmuEX0007427
- MmuEX0015603
- MmuEX0015605
- MmuEX0015607
- MmuEX0015608
- MmuEX0018961
- MmuEX0027619
- MmuEX0037682
- MmuEX0037683
- MmuEX0050063
- MmuEX1009506
- MmuEX6099491
- MmuEX6099559
- MmuINT0001915
- MmuINT0001917
- MmuINT0001920
- MmuINT0007787
- MmuINT0007798
- MmuINT0007805
- MmuINT0007827
- MmuINT0007831
- MmuINT0023114
- MmuINT0023115
- MmuINT0023116
- MmuINT0023117
- MmuINT0023118
- MmuINT0023119
- MmuINT0027152
- MmuINT0027154
- MmuINT0037298
- MmuINT0037299
- MmuINT0037300
- MmuINT0037301
- MmuINT0037302
- MmuINT0052962
- MmuINT0052963
- MmuINT0052966
- MmuINT0052967
- MmuINT0052968
- MmuINT0052969
- MmuINT0052971
- MmuINT0052972
- MmuINT0052973
- MmuINT0057672
- MmuINT0057675
- MmuINT0063572
- MmuINT0063573
- MmuINT0063577
- MmuINT0090240
- MmuINT0094770
- MmuINT0094771
- MmuINT0094772
- MmuINT0110053
- MmuINT0110058
- MmuINT0128480
- MmuINT0168759
- MmuINT1003301
- MmuINT1027719
Conservation
Fruitfly
(dm6)
No conservation detected
Primers PCR
Suggestions for RT-PCR validation
F:
ACAACACACACACACACACAAC
R:
ACCAGGAGAAGAAGGGGCG
Band lengths:
258-433
Functional annotations
There are 1 annotated functions for this event
PMID: 21725316
Encodes an experimentally validated Eukaryotic Linear Motif (ELM). Method: alanine scanning, coimmunoprecipitation, protein kinase assay, western blot. ELM ID: ELMI003387; ELM sequence: KDDSLLL; Overlap: FULL
GENOMIC CONTEXT[edit]
INCLUSION PATTERN[edit]
SPECIAL DATASETS
- Ribosome-engaged transcriptomes of neuronal types
- Neural differentiation time course
- Muscular differentiation time course
- Spermatogenesis cell types
- Reprogramming of fibroblasts to iPSCs
- Hematopoietic precursors and cell types